Working with slide-seq data¶
[1]:
import stlearn as st
import warnings
warnings.filterwarnings('ignore')
The data is downloaded from https://www.spatialomics.org/SpatialDB/download.php
Technique |
PMID |
Title |
Expression |
SV genes |
|---|---|---|---|---|
Slide-seq |
30923225 |
Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution |
slideseq_30923225.tar.gz |
slideseq_30923225_SVG.tar.gz |
Read Slide-seq data
[2]:
data = st.ReadSlideSeq(count_matrix_file="SlideSeq/slideseq_30923225/Puck_180413_7.count",
spatial_file="SlideSeq/slideseq_30923225/Puck_180413_7.idx")
[3]:
data
[3]:
AnnData object with n_obs × n_vars = 12282 × 3235
obs: 'index', 'imagecol', 'imagerow'
var: 'ENSEMBL'
uns: 'spatial'
obsm: 'spatial'
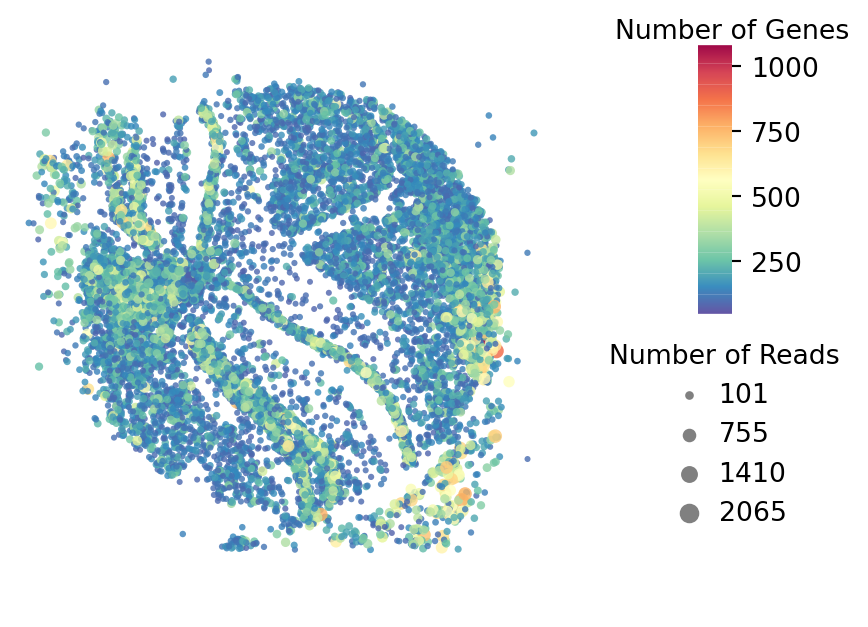
Plot for QC
[4]:
st.pl.QC_plot(data)
Preprocessing data
[5]:
st.pp.filter_genes(data,min_cells=3)
st.pp.normalize_total(data)
st.pp.log1p(data)
st.pp.scale(data)
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Scale step is finished in adata.X
Run PCA
[6]:
st.em.run_pca(data,n_comps=50,random_state=0)
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
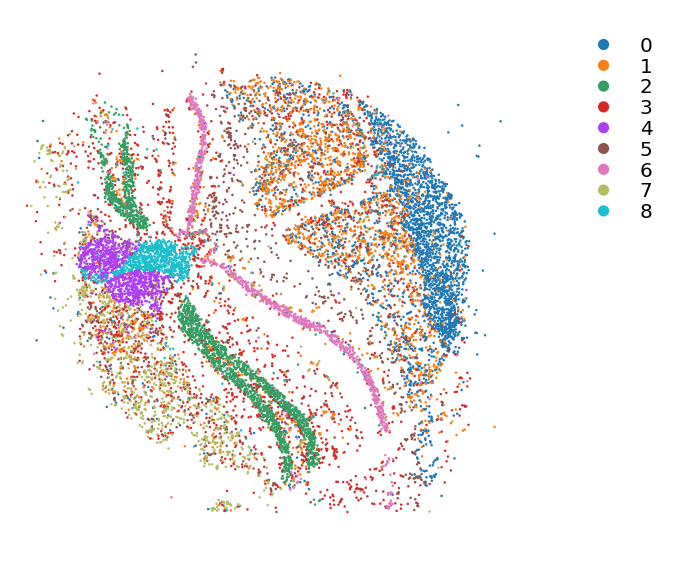
Run and plot louvain clustering
[7]:
st.pp.neighbors(data,n_neighbors=25,use_rep='X_pca',random_state=0)
Created k-Nearest-Neighbor graph in adata.uns['neighbors']
[8]:
st.tl.clustering.louvain(data,random_state=0)
Applying Louvain clustering ...
Louvain clustering is done! The labels are stored in adata.obs['louvain']
[9]:
st.pl.cluster_plot(data,use_label="louvain",tissue_alpha=1,spot_size=1,show_legend=True)