stSME clustering tutorial¶
stSME is a novel normalisation method implemented in stLearn software.
It’s designed for spatial transcriptomics data and utilised tissue Spatial location, Morphology, , and gene Expression.
This tutorial demonstrates how to use stLearn to perform stSME clustering for spatial transcriptomics data
In this tutorial we first focus on Mouse Brain (Coronal) Visium dataset from 10x genomics website.
Mouse Brain (Coronal)¶
1. Preparation¶
[1]:
# import module
import stlearn as st
from pathlib import Path
st.settings.set_figure_params(dpi=180)
[2]:
# specify PATH to data
BASE_PATH = Path("/home/uqysun19/60days/10x_visium/mouse_brain_coronal")
# spot tile is the intermediate result of image pre-processing
TILE_PATH = Path("/tmp/tiles")
TILE_PATH.mkdir(parents=True, exist_ok=True)
# output path
OUT_PATH = Path("/home/uqysun19/60days/stlearn_plot/mouse_brain_coronl")
OUT_PATH.mkdir(parents=True, exist_ok=True)
[3]:
# load data
data = st.Read10X(BASE_PATH)
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
[4]:
# pre-processing for gene count table
st.pp.filter_genes(data,min_cells=1)
st.pp.normalize_total(data)
st.pp.log1p(data)
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
[5]:
# pre-processing for spot image
st.pp.tiling(data, TILE_PATH)
# this step uses deep learning model to extract high-level features from tile images
# may need few minutes to be completed
st.pp.extract_feature(data)
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
2. run stSME clustering¶
[6]:
# run PCA for gene expression data
st.em.run_pca(data,n_comps=50)
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
[7]:
data_SME = data.copy()
# apply stSME to normalise log transformed data
st.spatial.SME.SME_normalize(data_SME, use_data="raw")
data_SME.X = data_SME.obsm['raw_SME_normalized']
st.pp.scale(data_SME)
st.em.run_pca(data_SME,n_comps=50)
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
[8]:
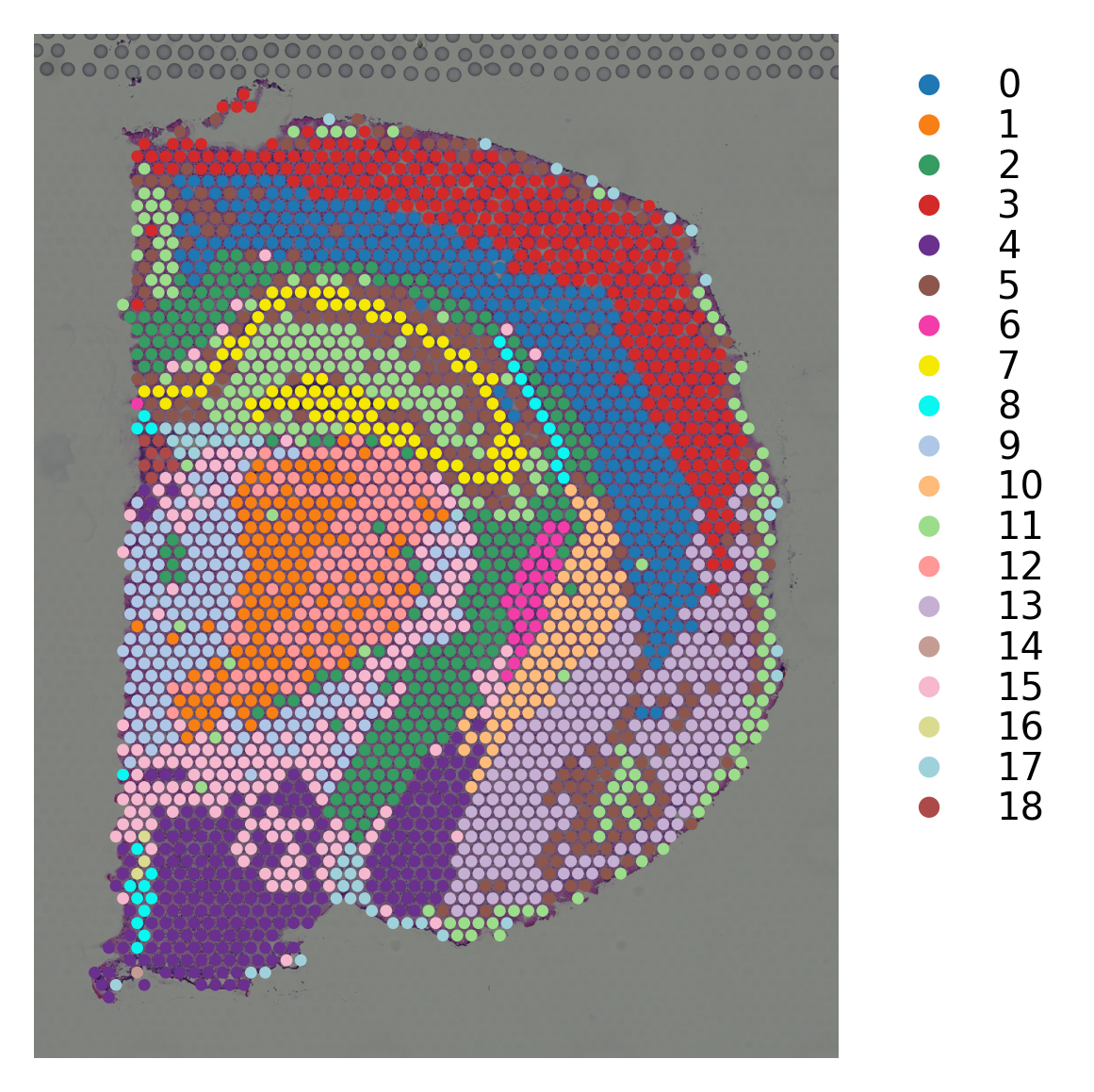
# K-means clustering on stSME normalised PCA
st.tl.clustering.kmeans(data_SME,n_clusters=19, use_data="X_pca", key_added="X_pca_kmeans")
st.pl.cluster_plot(data_SME, use_label="X_pca_kmeans")
Applying Kmeans clustering ...
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
[9]:
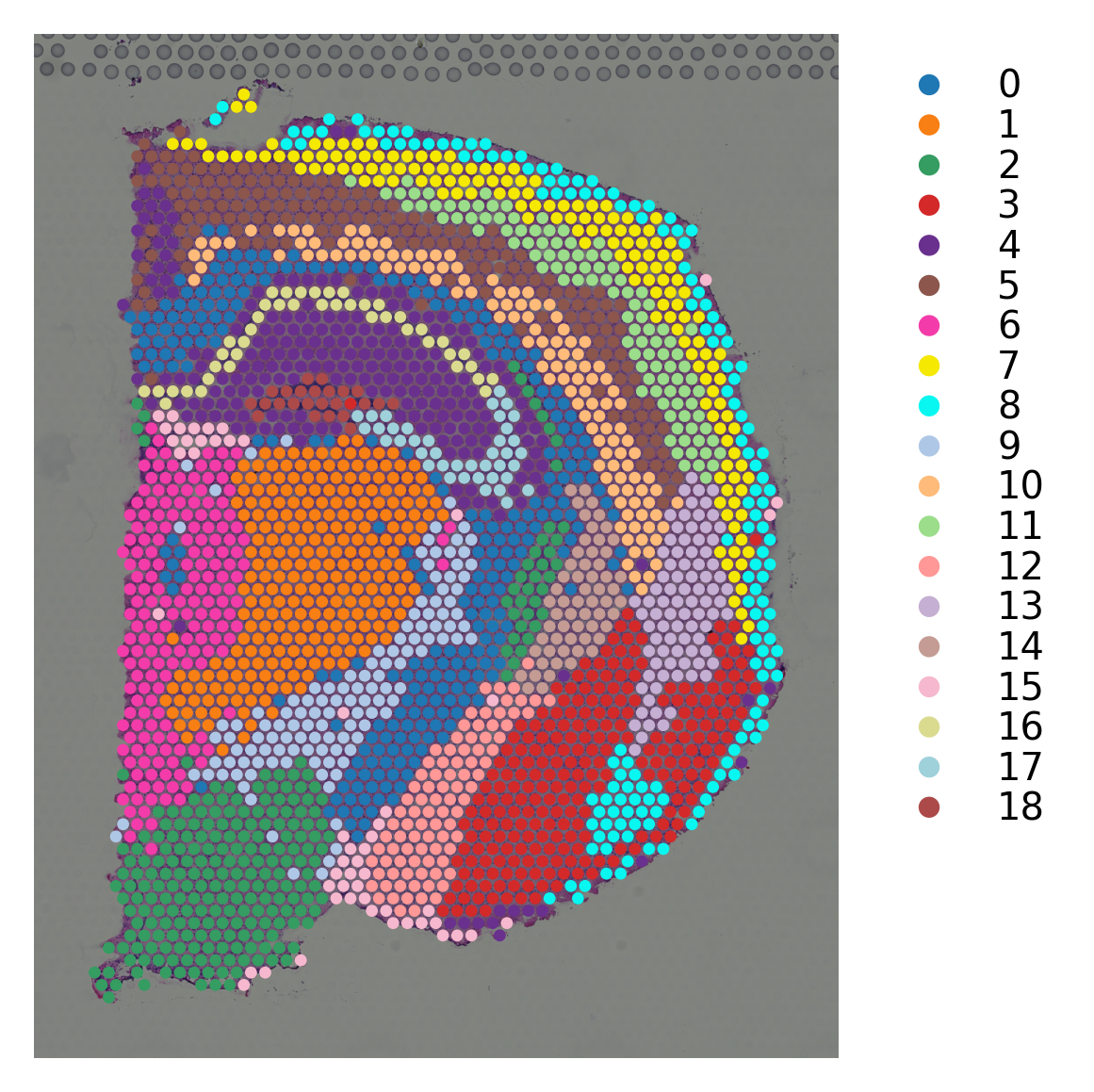
# louvain clustering on stSME normalised data
st.pp.neighbors(data_SME,n_neighbors=17,use_rep='X_pca')
st.tl.clustering.louvain(data_SME, resolution=1.19)
st.pl.cluster_plot(data_SME,use_label="louvain")
Created k-Nearest-Neighbor graph in adata.uns['neighbors']
Applying Louvain clustering ...
Louvain clustering is done! The labels are stored in adata.obs['louvain']
we now move to Mouse Brain (Sagittal Posterior) Visium dataset from 10x genomics website.
Mouse Brain (Sagittal Posterior)¶
1. Preparation¶
[10]:
# specify PATH to data
BASE_PATH = Path("/home/uqysun19/60days/10x_visium/mouse_brain_s_p_1/")
# spot tile is the intermediate result of image pre-processing
TILE_PATH = Path("/tmp/tiles")
TILE_PATH.mkdir(parents=True, exist_ok=True)
# outpot path
OUT_PATH = Path("/home/uqysun19/60days/stlearn_plot/mouse_brain_s_p_1/")
OUT_PATH.mkdir(parents=True, exist_ok=True)
[11]:
# load data
data = st.Read10X(BASE_PATH)
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
[12]:
# pre-processing for gene count table
st.pp.filter_genes(data,min_cells=1)
st.pp.normalize_total(data)
st.pp.log1p(data)
st.pp.scale(data)
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Scale step is finished in adata.X
[13]:
# pre-processing for spot image
st.pp.tiling(data, TILE_PATH)
# this step uses deep learning model to extract high-level features from tile images
# may need few minutes to be completed
st.pp.extract_feature(data)
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
2. run stSME clustering¶
[14]:
# run PCA for gene expression data
st.em.run_pca(data,n_comps=50)
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
[15]:
data_SME = data.copy()
# apply stSME to normalise log transformed data
# with weights from morphological Similarly and physcial distance
st.spatial.SME.SME_normalize(data_SME, use_data="raw",
weights="weights_matrix_pd_md")
data_SME.X = data_SME.obsm['raw_SME_normalized']
st.pp.scale(data_SME)
st.em.run_pca(data_SME,n_comps=50)
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
[16]:
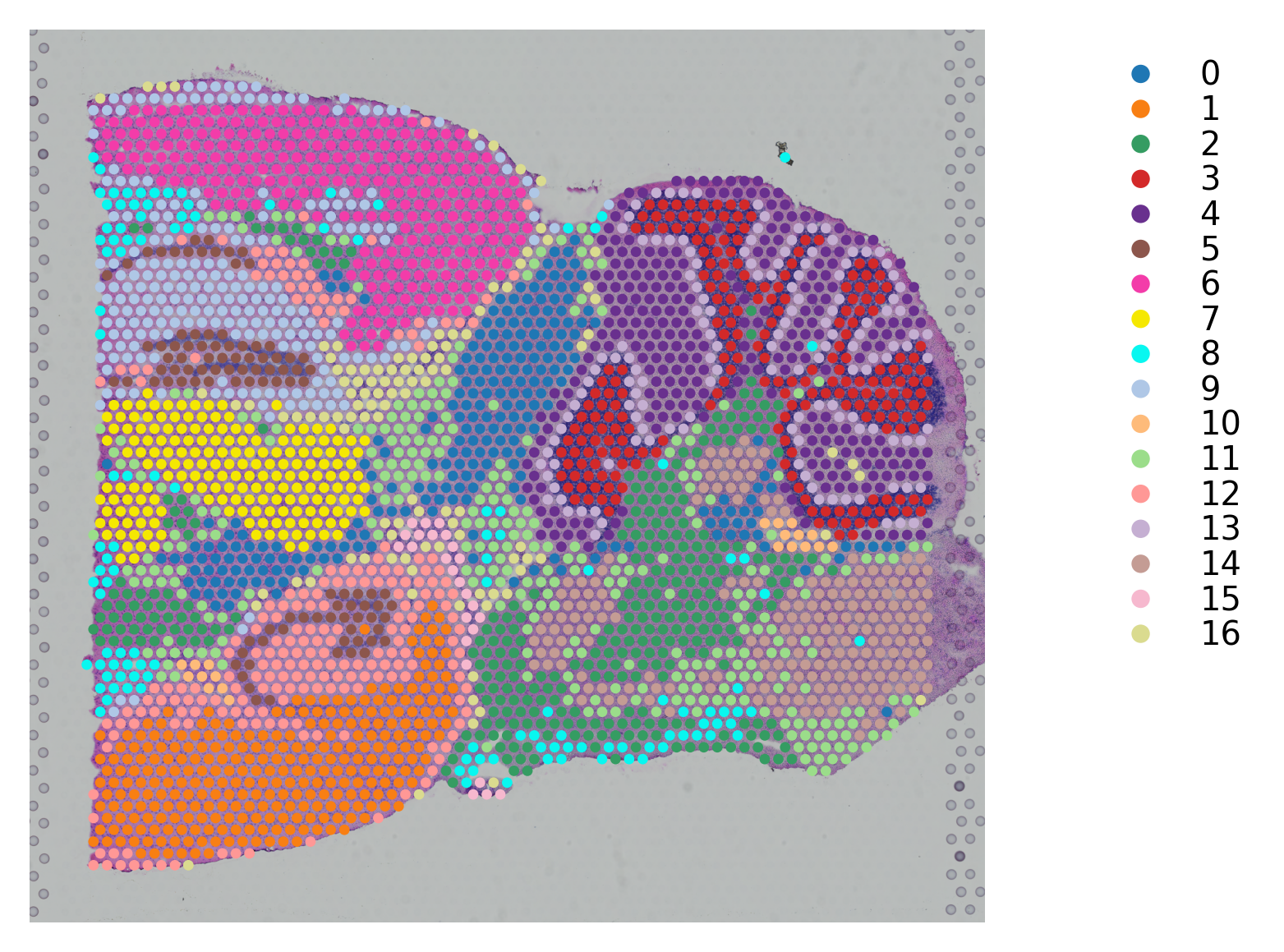
# K-means clustering on stSME normalised PCA
st.tl.clustering.kmeans(data_SME,n_clusters=17, use_data="X_pca", key_added="X_pca_kmeans")
st.pl.cluster_plot(data_SME, use_label="X_pca_kmeans")
Applying Kmeans clustering ...
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
[17]:
# louvain clustering on stSME normalised data
st.pp.neighbors(data_SME,n_neighbors=20,use_rep='X_pca')
st.tl.clustering.louvain(data_SME)
st.pl.cluster_plot(data_SME,use_label="louvain")
Created k-Nearest-Neighbor graph in adata.uns['neighbors']
Applying Louvain clustering ...
Louvain clustering is done! The labels are stored in adata.obs['louvain']

Then we apply stSME clustering on Human Brain dorsolateral prefrontal cortex (DLPFC) Visium dataset from this paper.
Human Brain dorsolateral prefrontal cortex (DLPFC)¶
[18]:
import pandas as pd
from sklearn.metrics import adjusted_rand_score, normalized_mutual_info_score, silhouette_score, \
homogeneity_completeness_v_measure
from sklearn.metrics.cluster import contingency_matrix
from sklearn.preprocessing import LabelEncoder
import numpy as np
import scanpy
[19]:
def purity_score(y_true, y_pred):
# compute contingency matrix (also called confusion matrix)
cm = contingency_matrix(y_true, y_pred)
# return purity
return np.sum(np.amax(cm, axis=0)) / np.sum(cm)
[20]:
# specify PATH to data
BASE_PATH = Path("/home/uqysun19/60days/Human_Brain_spatialLIBD")
[21]:
# here we include all 12 samples
sample_list = ["151507", "151508", "151509",
"151510", "151669", "151670",
"151671", "151672", "151673",
"151674", "151675", "151676"]
Ground truth¶
[22]:
for i in range(len(sample_list)):
sample = sample_list[i]
GROUND_TRUTH_PATH = BASE_PATH / sample / "cluster_labels_{}.csv".format(sample)
ground_truth_df = pd.read_csv(GROUND_TRUTH_PATH, sep=',', index_col=0)
ground_truth_df.index = ground_truth_df.index.map(lambda x: x[7:])
le = LabelEncoder()
ground_truth_le = le.fit_transform(list(ground_truth_df["ground_truth"].values))
ground_truth_df["ground_truth_le"] = ground_truth_le
# load data
data = st.Read10X(BASE_PATH / sample)
ground_truth_df = ground_truth_df.reindex(data.obs_names)
data.obs["ground_truth"] = pd.Categorical(ground_truth_df["ground_truth"])
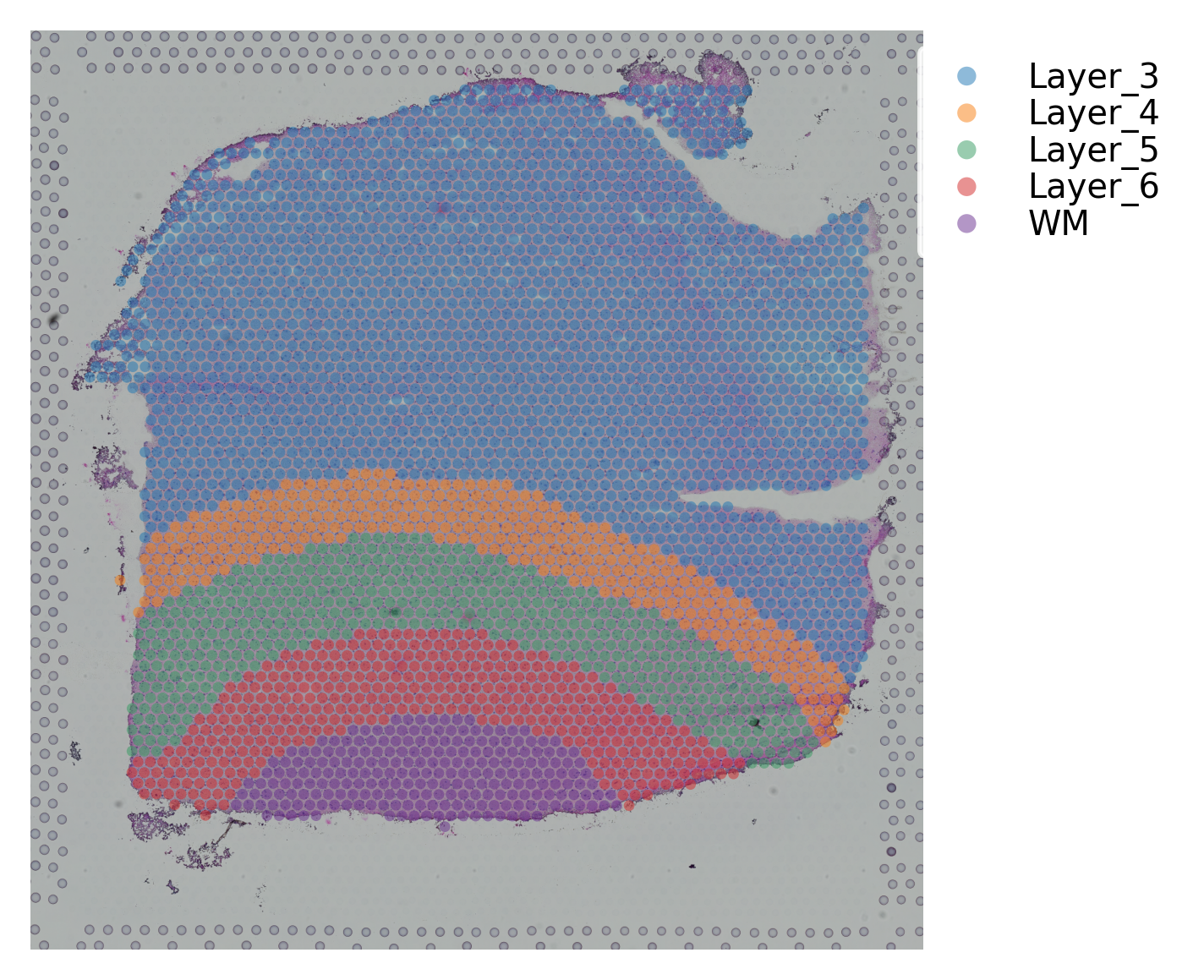
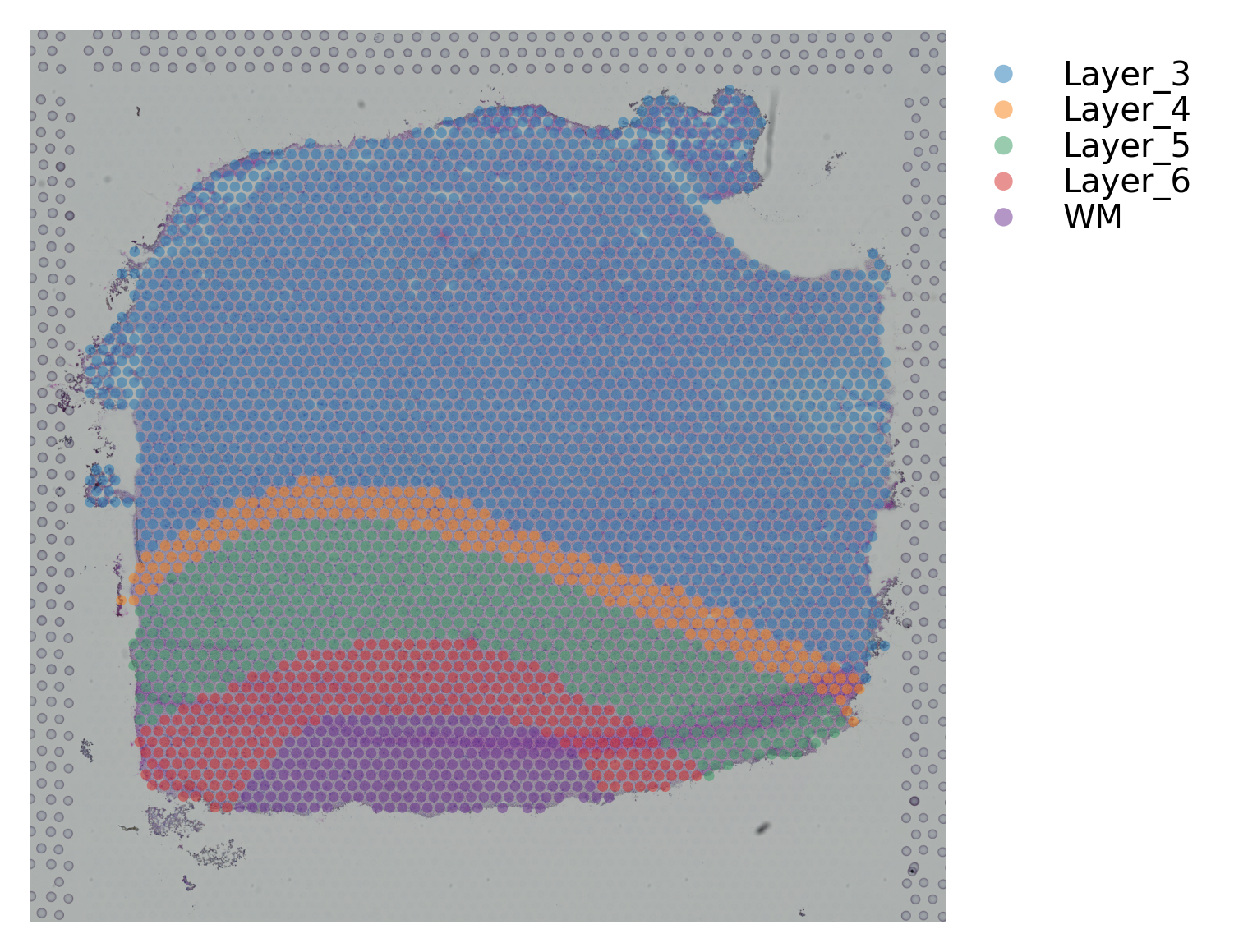
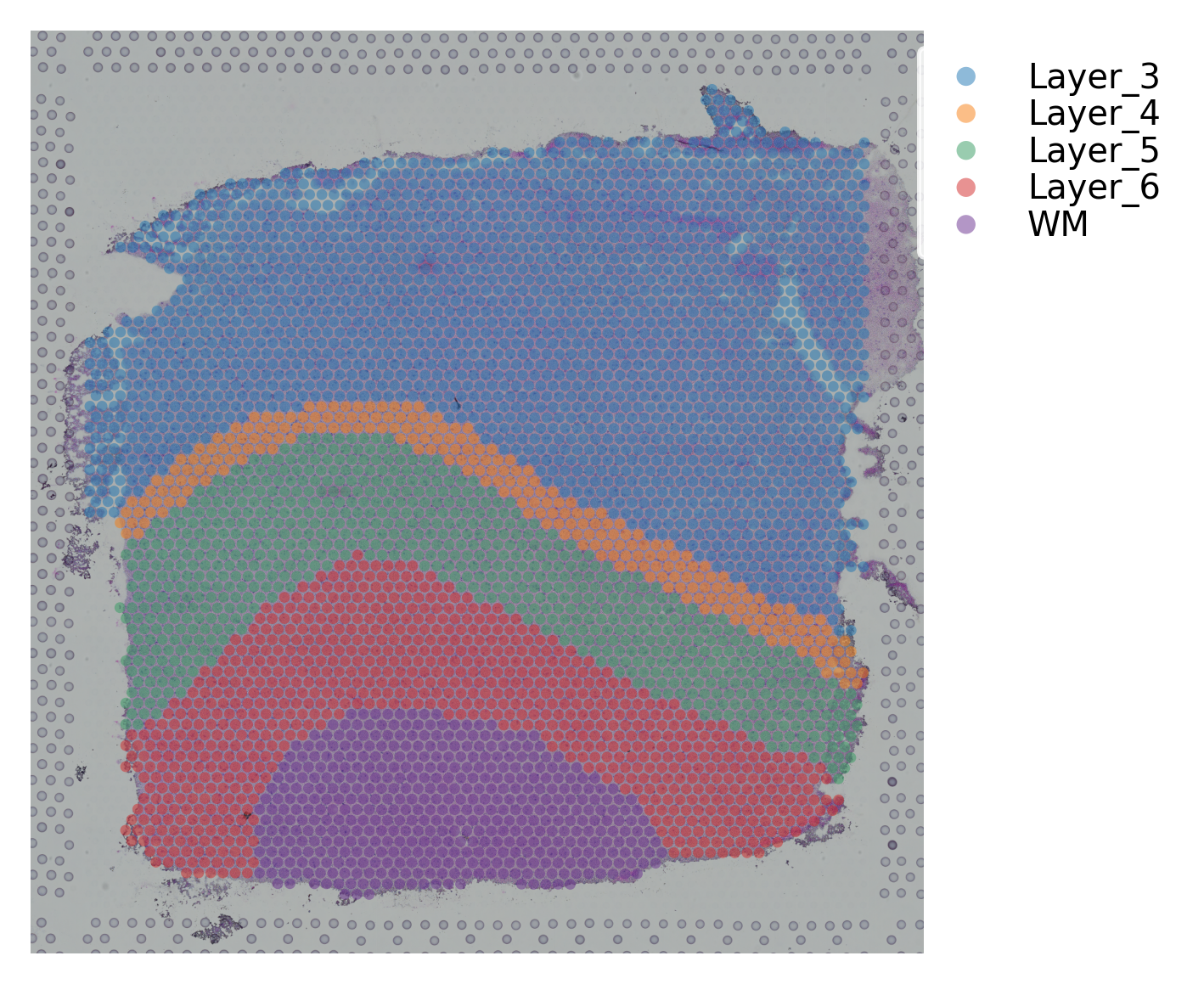
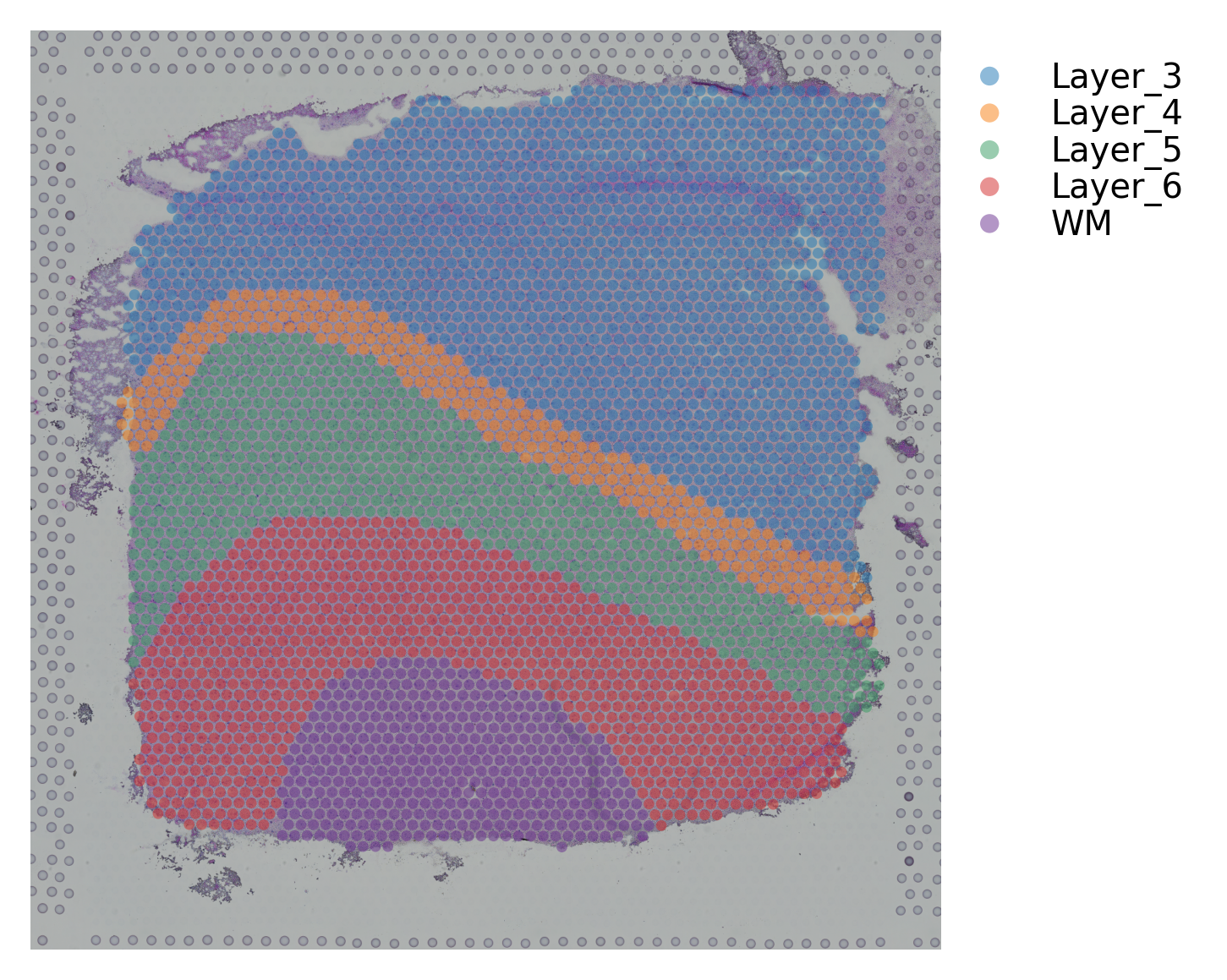
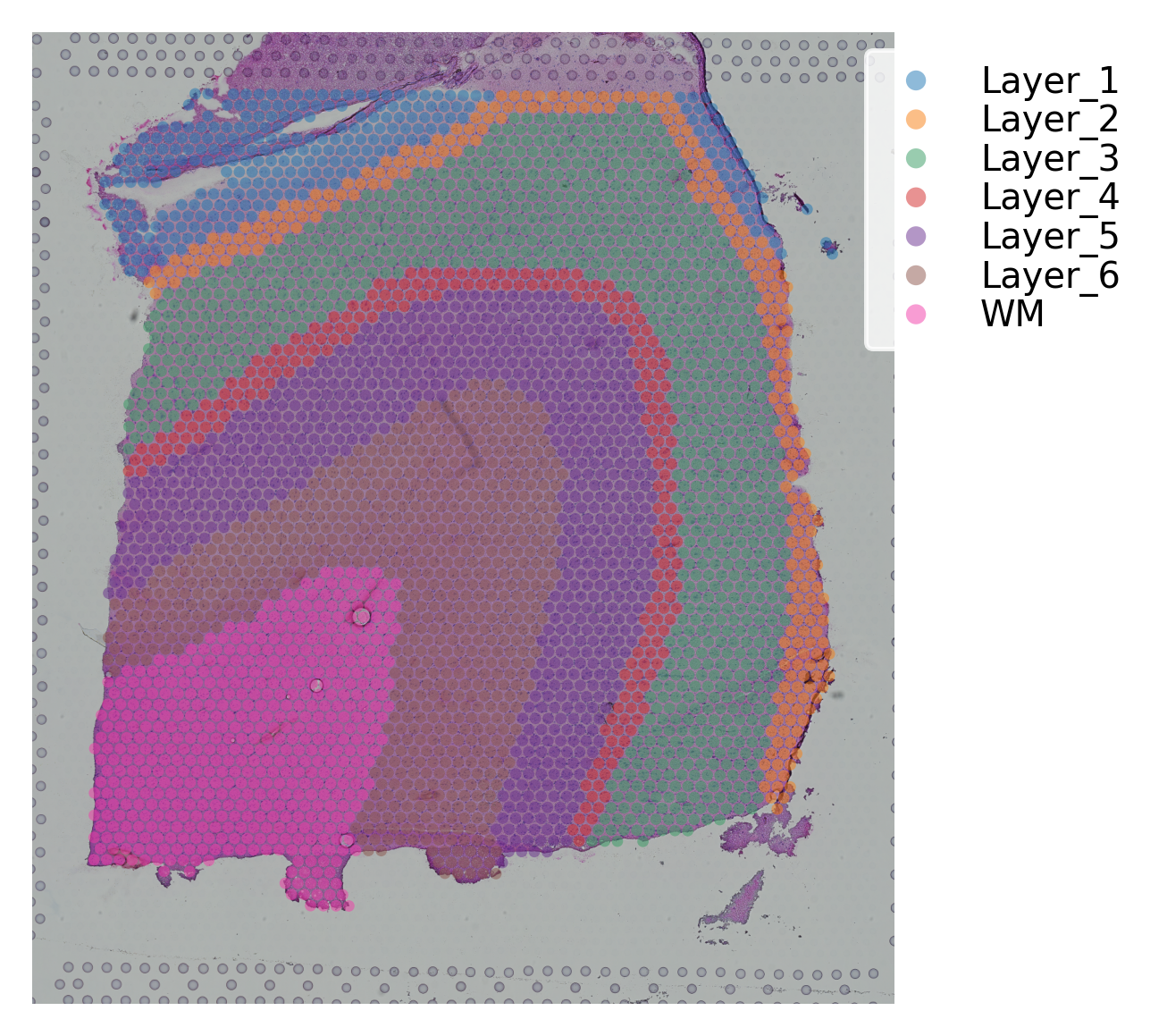
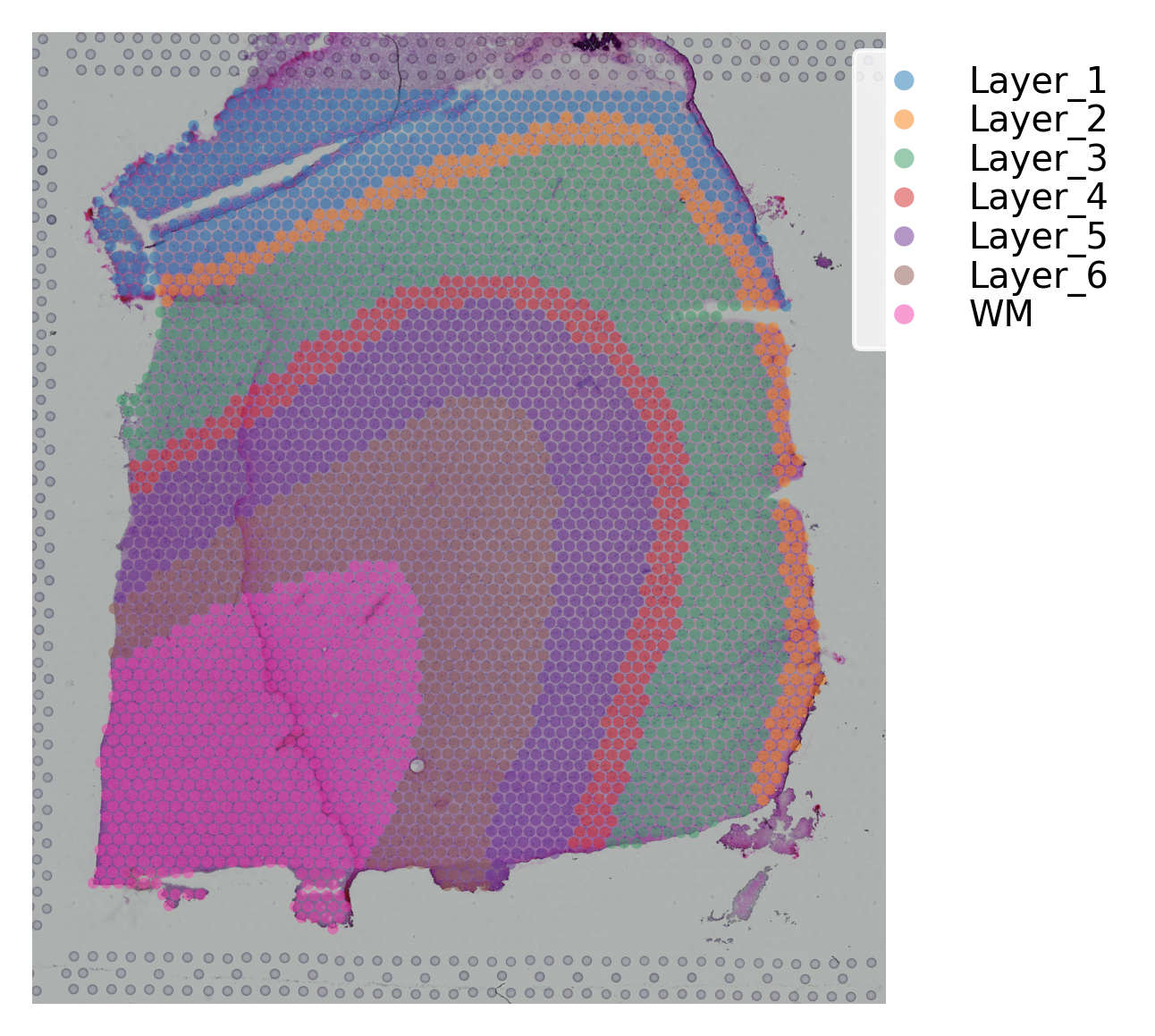
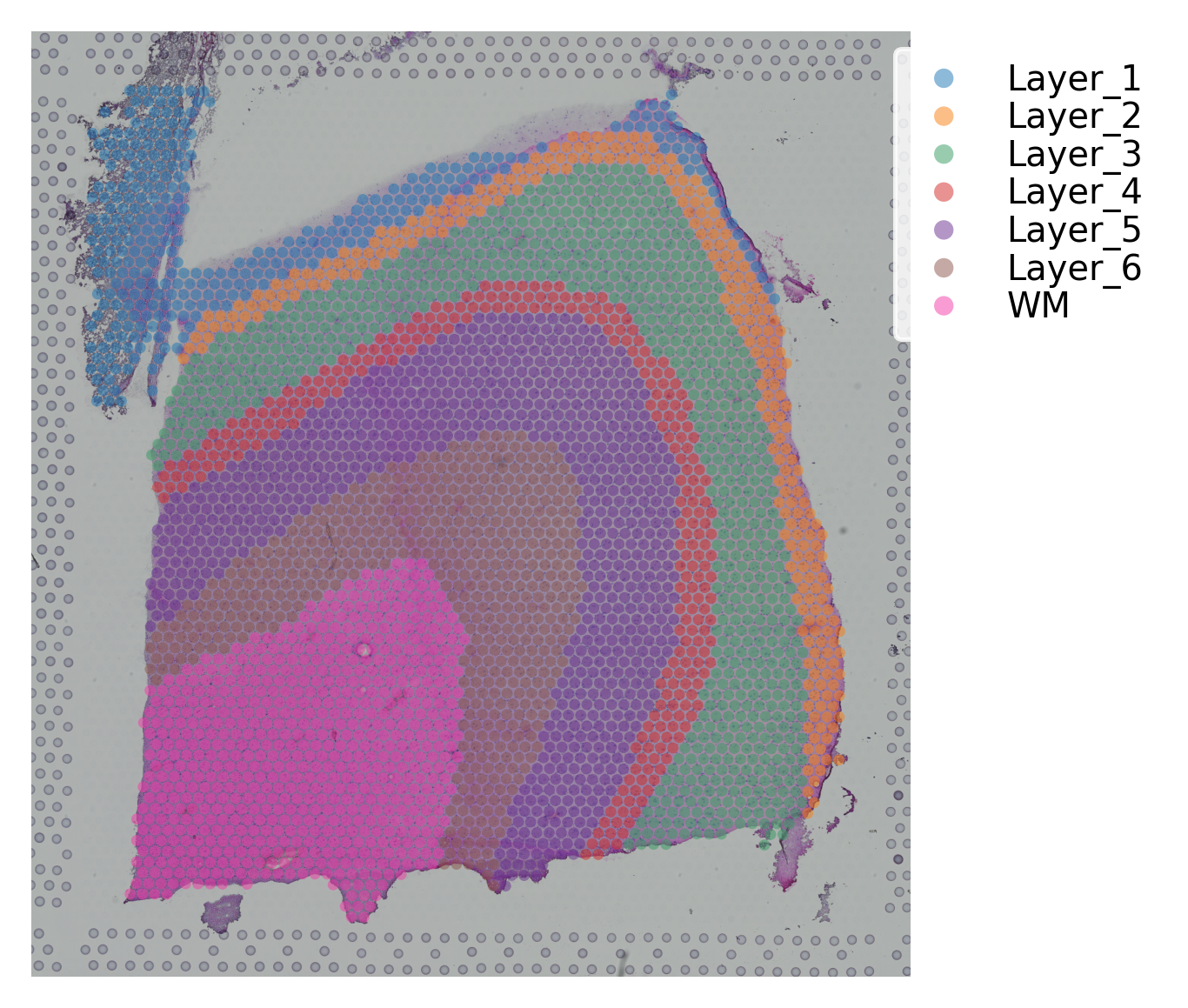
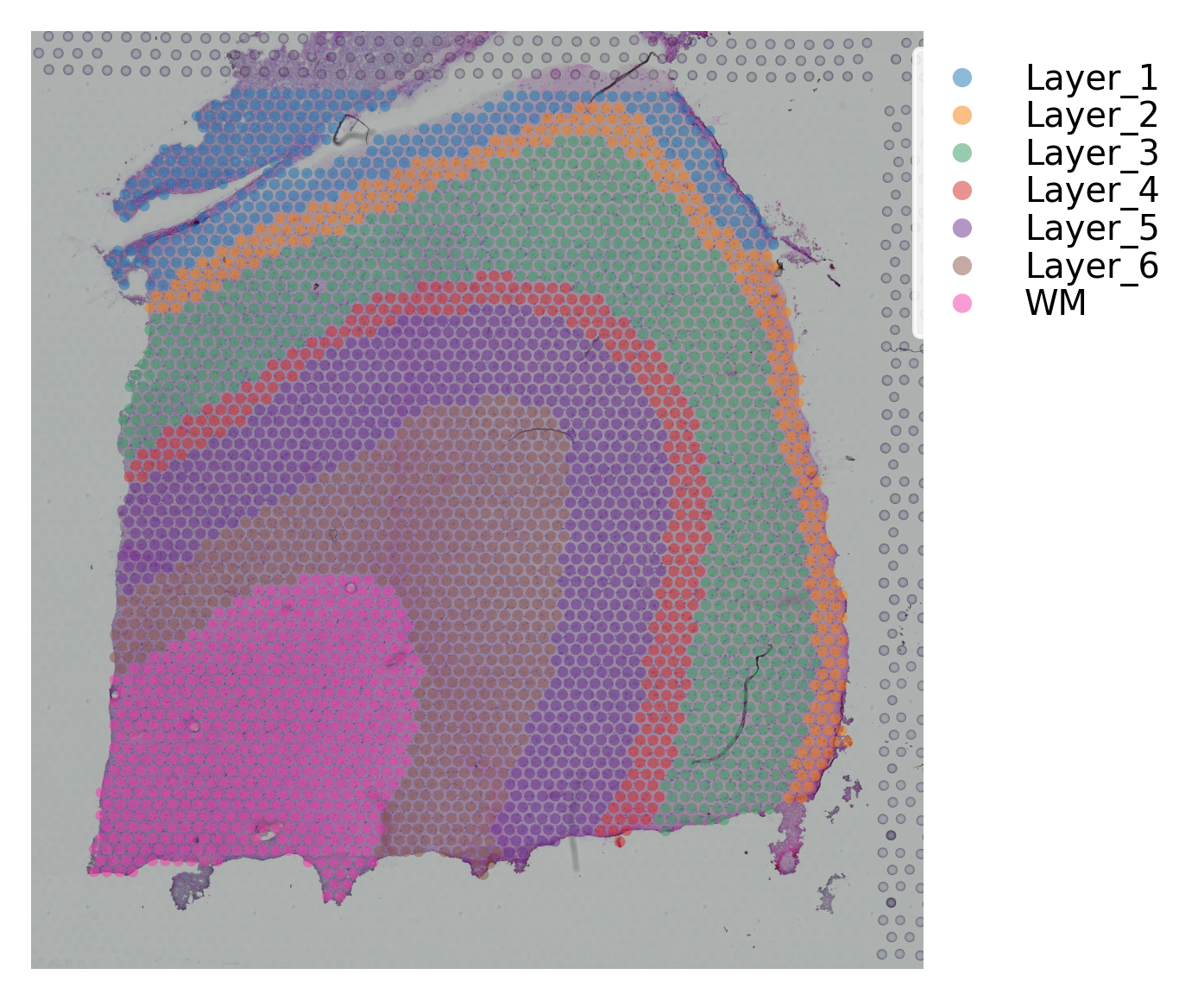
st.pl.cluster_plot(data, use_label="ground_truth", cell_alpha=0.5)
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
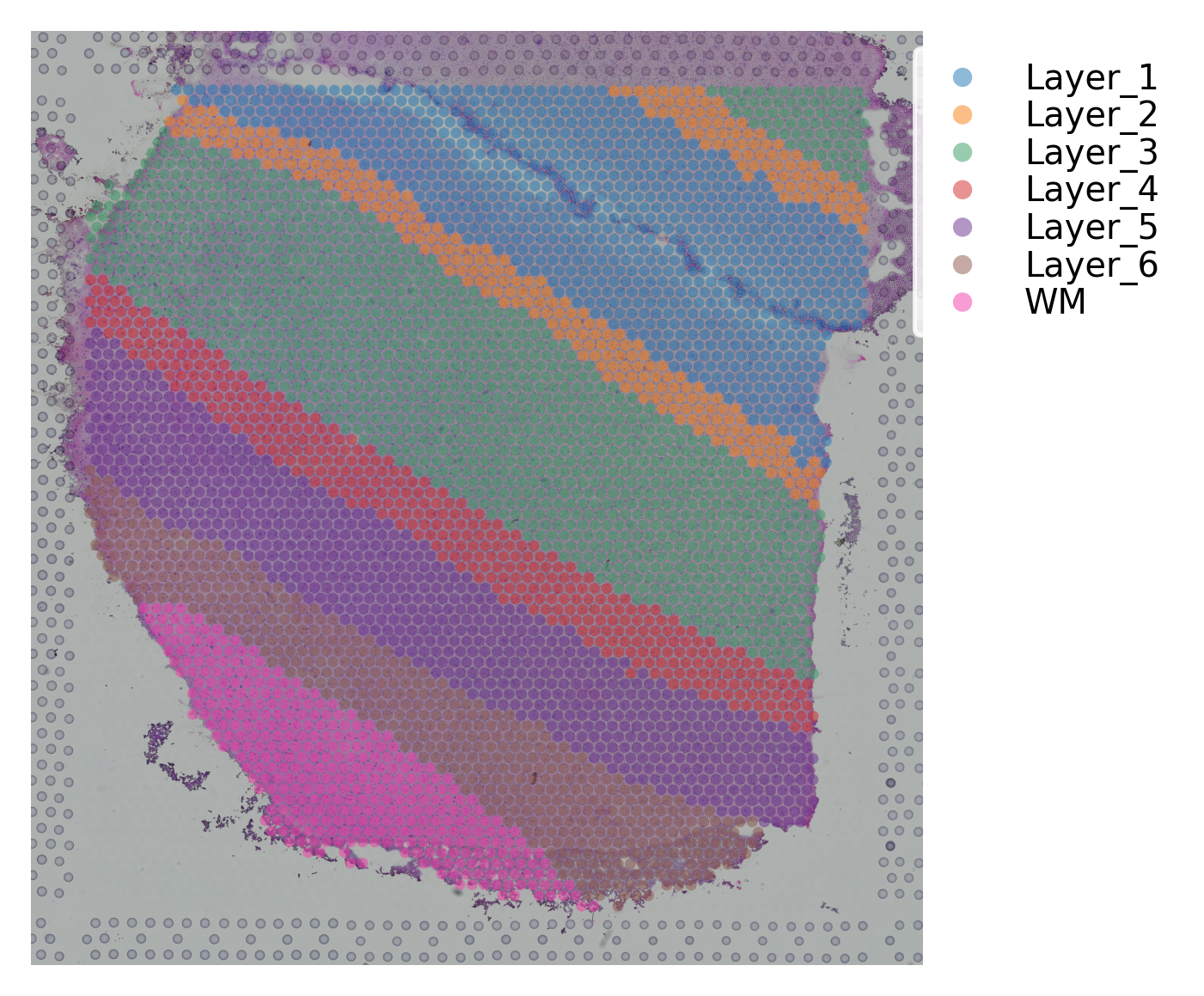
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
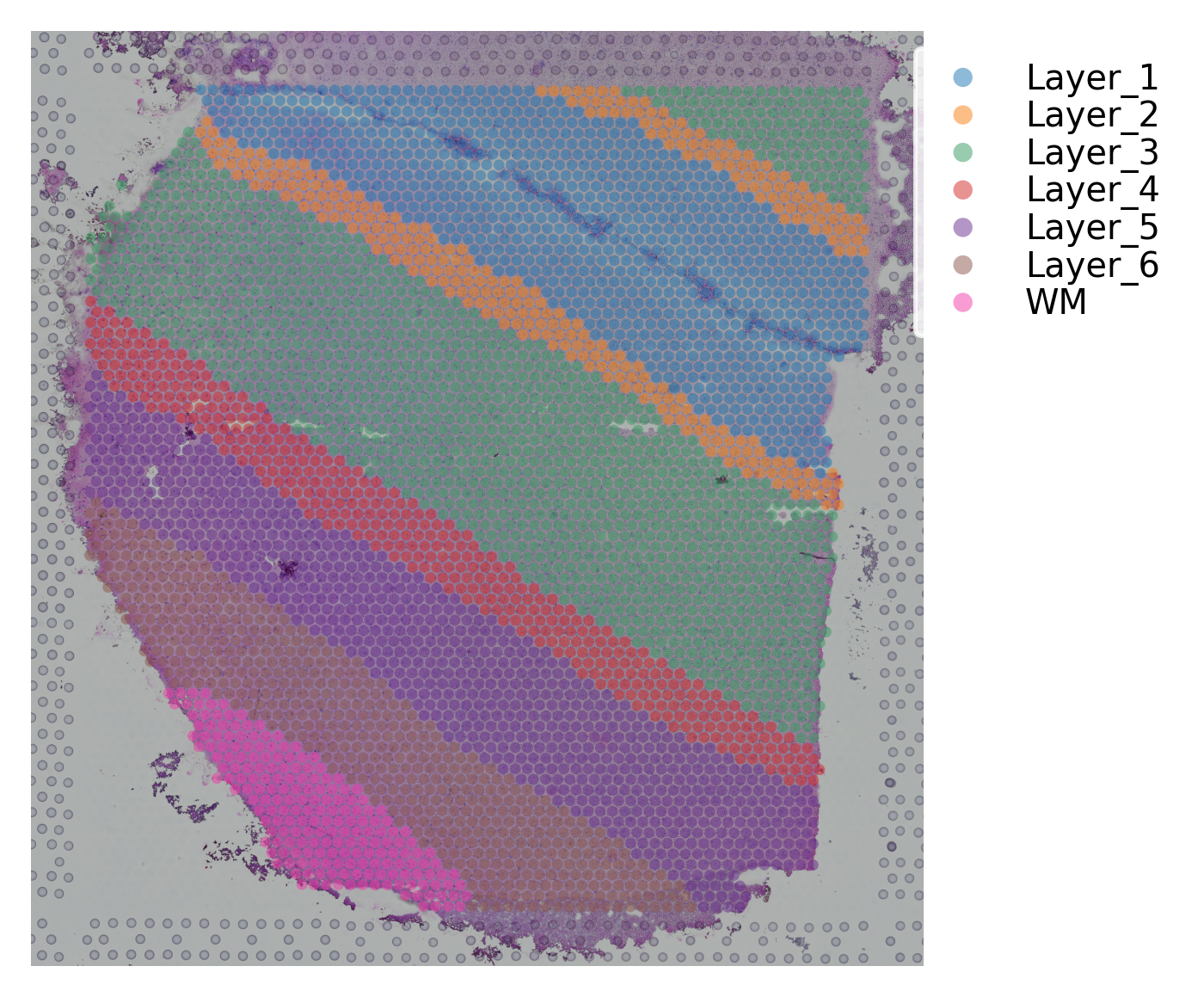
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
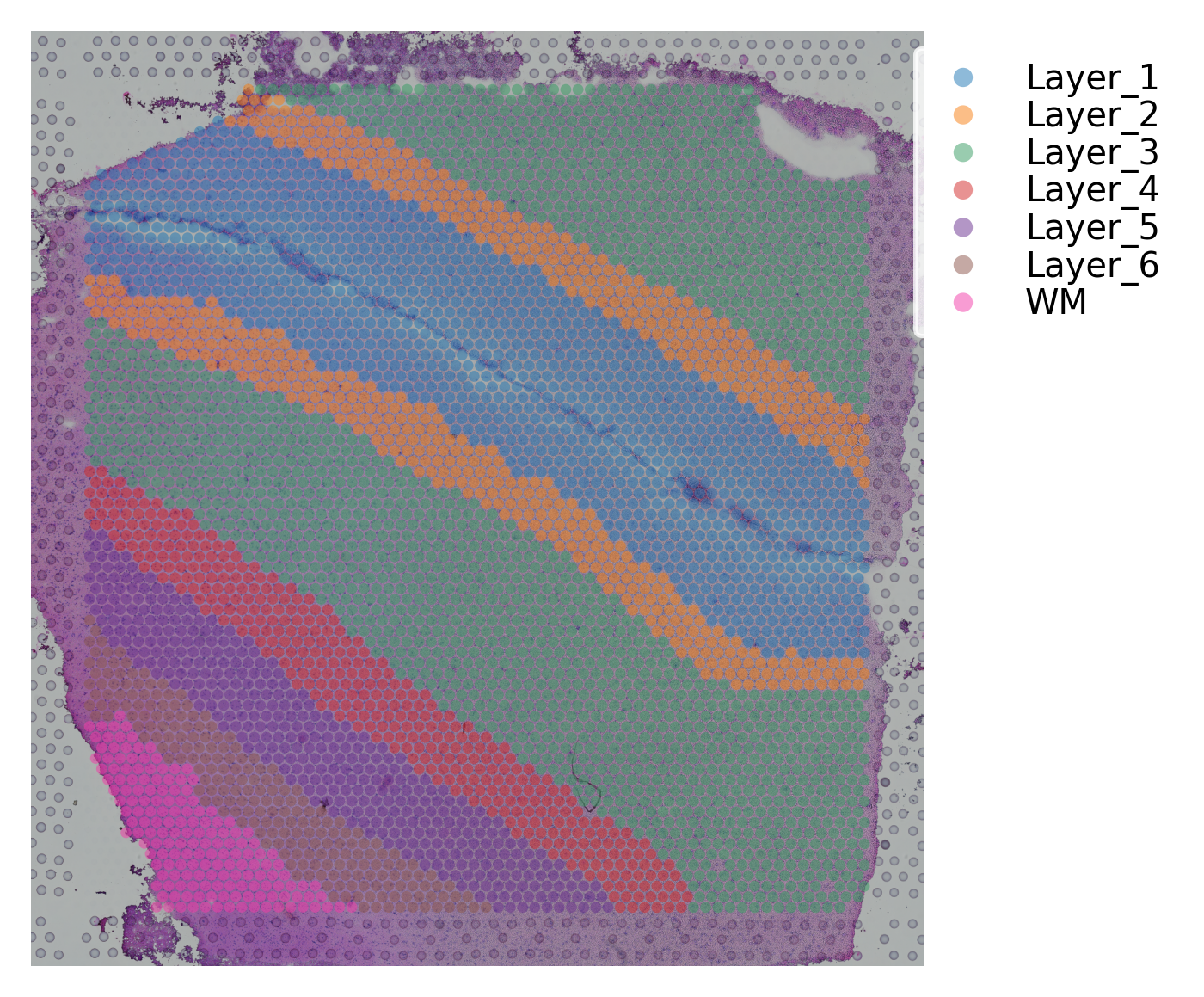
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
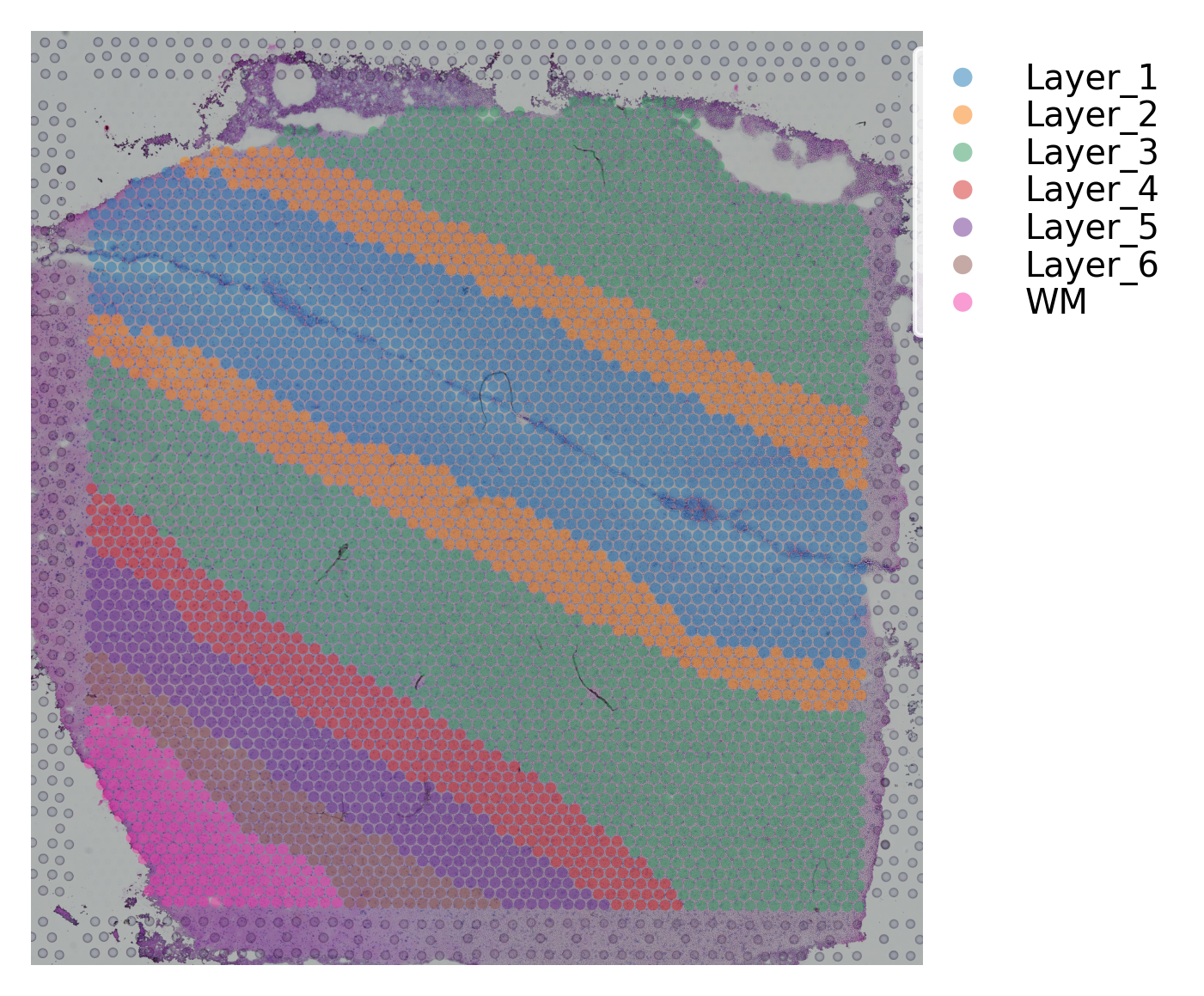
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
[23]:
def calculate_clustering_matrix(pred, gt, sample, methods_):
df = pd.DataFrame(columns=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"])
pca_ari = adjusted_rand_score(pred, gt)
df = df.append(pd.Series([sample, pca_ari, "pca", methods_, "Adjusted_Rand_Score"],
index=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"]), ignore_index=True)
pca_nmi = normalized_mutual_info_score(pred, gt)
df = df.append(pd.Series([sample, pca_nmi, "pca", methods_, "Normalized_Mutual_Info_Score"],
index=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"]), ignore_index=True)
pca_purity = purity_score(pred, gt)
df = df.append(pd.Series([sample, pca_purity, "pca", methods_, "Purity_Score"],
index=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"]), ignore_index=True)
pca_homogeneity, pca_completeness, pca_v_measure = homogeneity_completeness_v_measure(pred, gt)
df = df.append(pd.Series([sample, pca_homogeneity, "pca", methods_, "Homogeneity_Score"],
index=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"]), ignore_index=True)
df = df.append(pd.Series([sample, pca_completeness, "pca", methods_, "Completeness_Score"],
index=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"]), ignore_index=True)
df = df.append(pd.Series([sample, pca_v_measure, "pca", methods_, "V_Measure_Score"],
index=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"]), ignore_index=True)
return df
[24]:
df = pd.DataFrame(columns=['Sample', 'Score', 'PCA_or_UMAP', 'Method', "test"])
[25]:
for i in range(12):
sample = sample_list[i]
GROUND_TRUTH_PATH = BASE_PATH / sample / "cluster_labels_{}.csv".format(sample)
ground_truth_df = pd.read_csv(GROUND_TRUTH_PATH, sep=',', index_col=0)
ground_truth_df.index = ground_truth_df.index.map(lambda x: x[7:])
le = LabelEncoder()
ground_truth_le = le.fit_transform(list(ground_truth_df["ground_truth"].values))
ground_truth_df["ground_truth_le"] = ground_truth_le
TILE_PATH = Path("/tmp/{}_tiles".format(sample))
TILE_PATH.mkdir(parents=True, exist_ok=True)
data = st.Read10X(BASE_PATH / sample)
ground_truth_df = ground_truth_df.reindex(data.obs_names)
n_cluster = len((set(ground_truth_df["ground_truth"]))) - 1
data.obs['ground_truth'] = ground_truth_df["ground_truth"]
ground_truth_le = ground_truth_df["ground_truth_le"]
# pre-processing for gene count table
st.pp.filter_genes(data,min_cells=1)
st.pp.normalize_total(data)
st.pp.log1p(data)
# run PCA for gene expression data
st.em.run_pca(data,n_comps=15)
# pre-processing for spot image
st.pp.tiling(data, TILE_PATH)
# this step uses deep learning model to extract high-level features from tile images
# may need few minutes to be completed
st.pp.extract_feature(data)
# stSME
st.spatial.SME.SME_normalize(data, use_data="raw", weights="physical_distance")
data_ = data.copy()
data_.X = data_.obsm['raw_SME_normalized']
st.pp.scale(data_)
st.em.run_pca(data_,n_comps=15)
st.tl.clustering.kmeans(data_, n_clusters=n_cluster, use_data="X_pca", key_added="X_pca_kmeans")
st.pl.cluster_plot(data_, use_label="X_pca_kmeans")
methods_ = "stSME_disk"
results_df = calculate_clustering_matrix(data_.obs["X_pca_kmeans"], ground_truth_le, sample, methods_)
df = df.append(results_df, ignore_index=True)
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
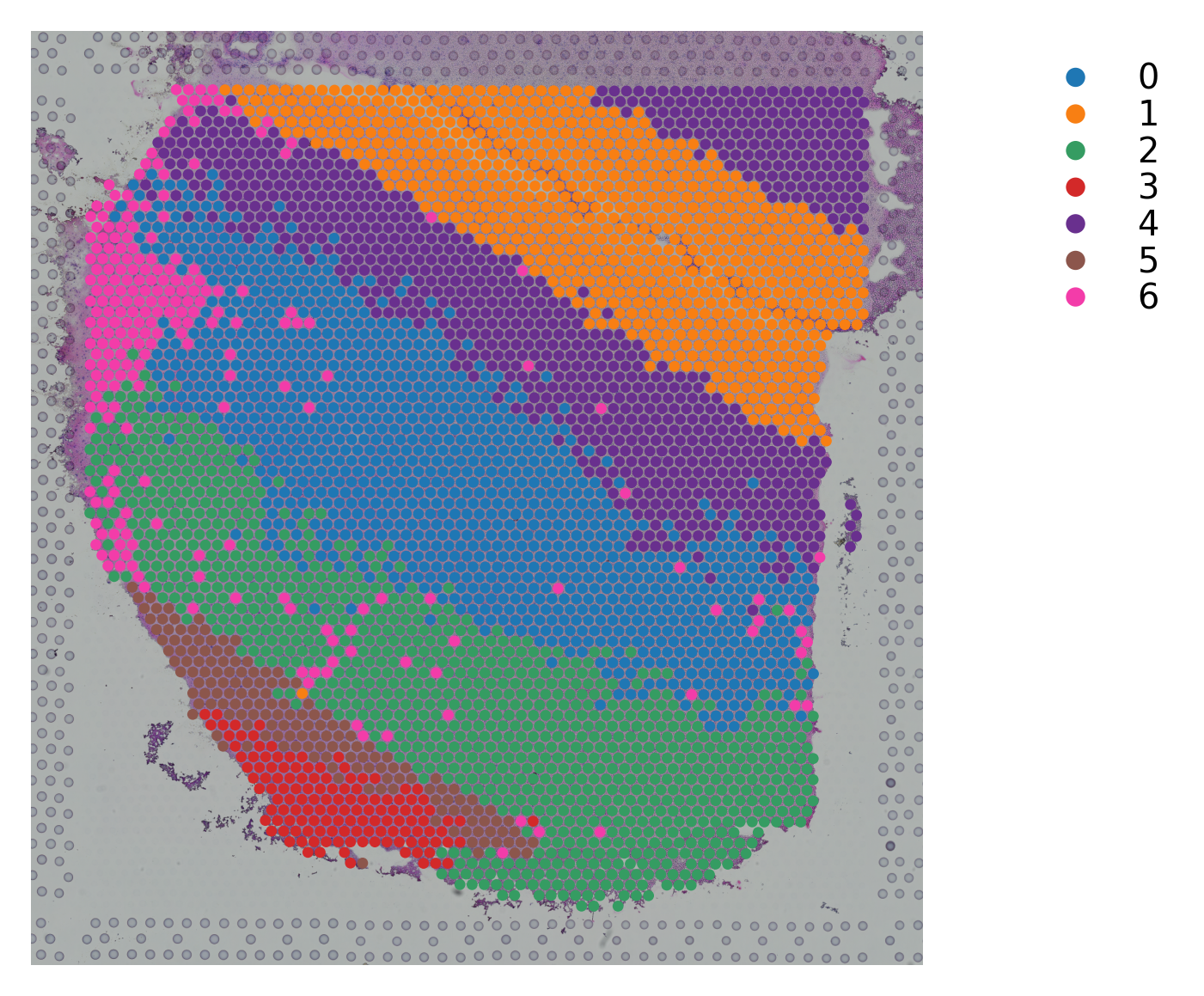
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:05 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
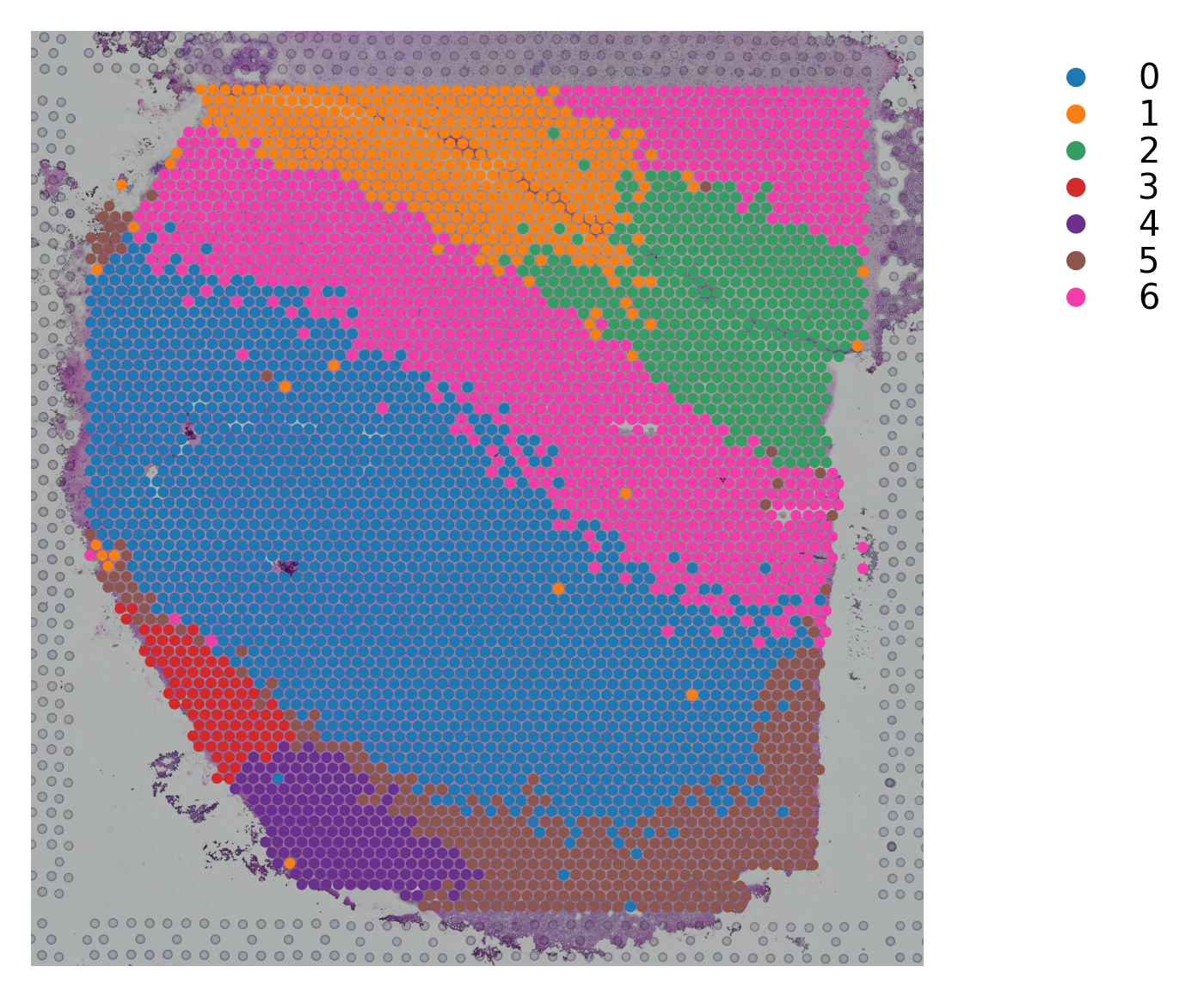
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:05 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
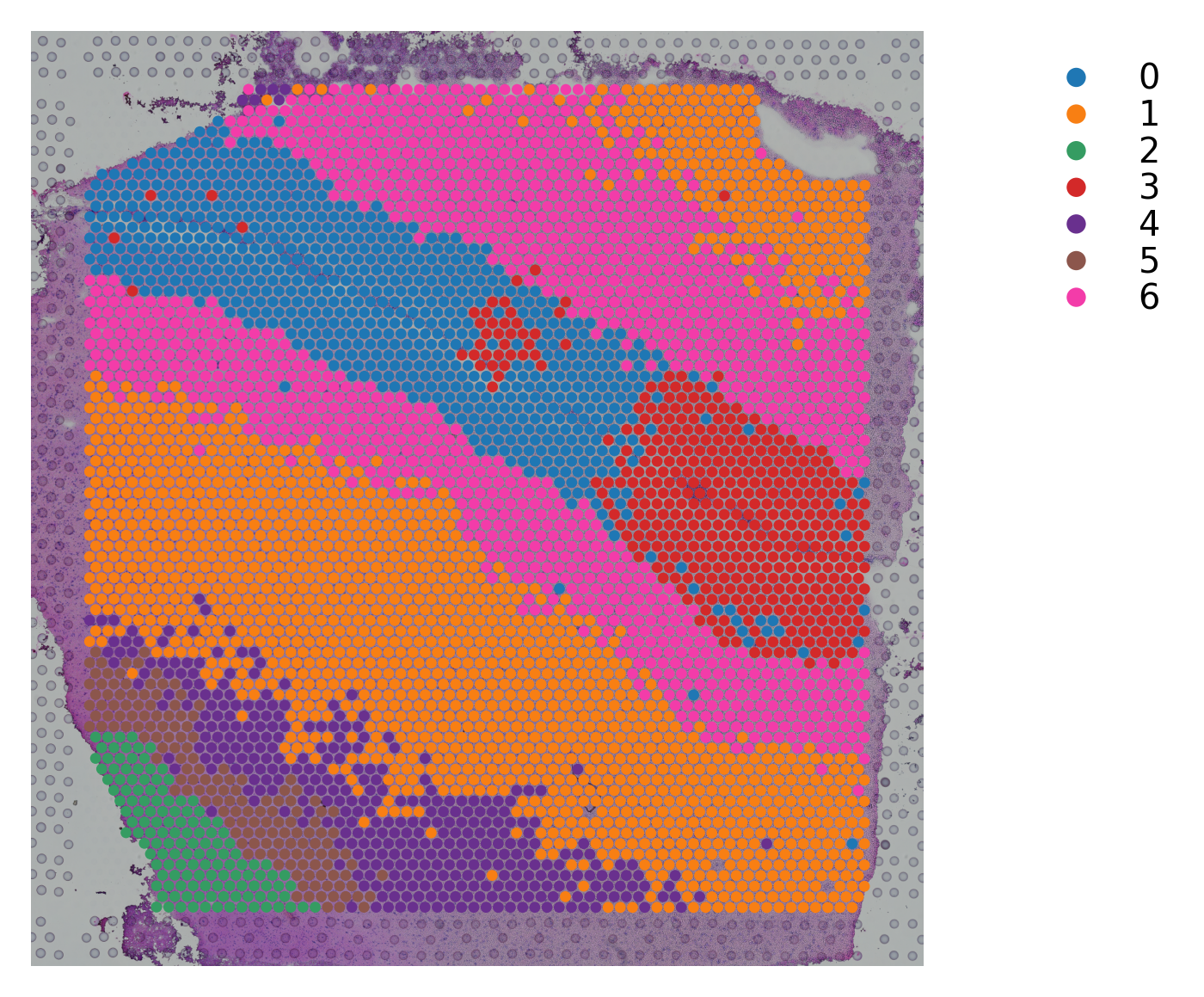
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:05 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
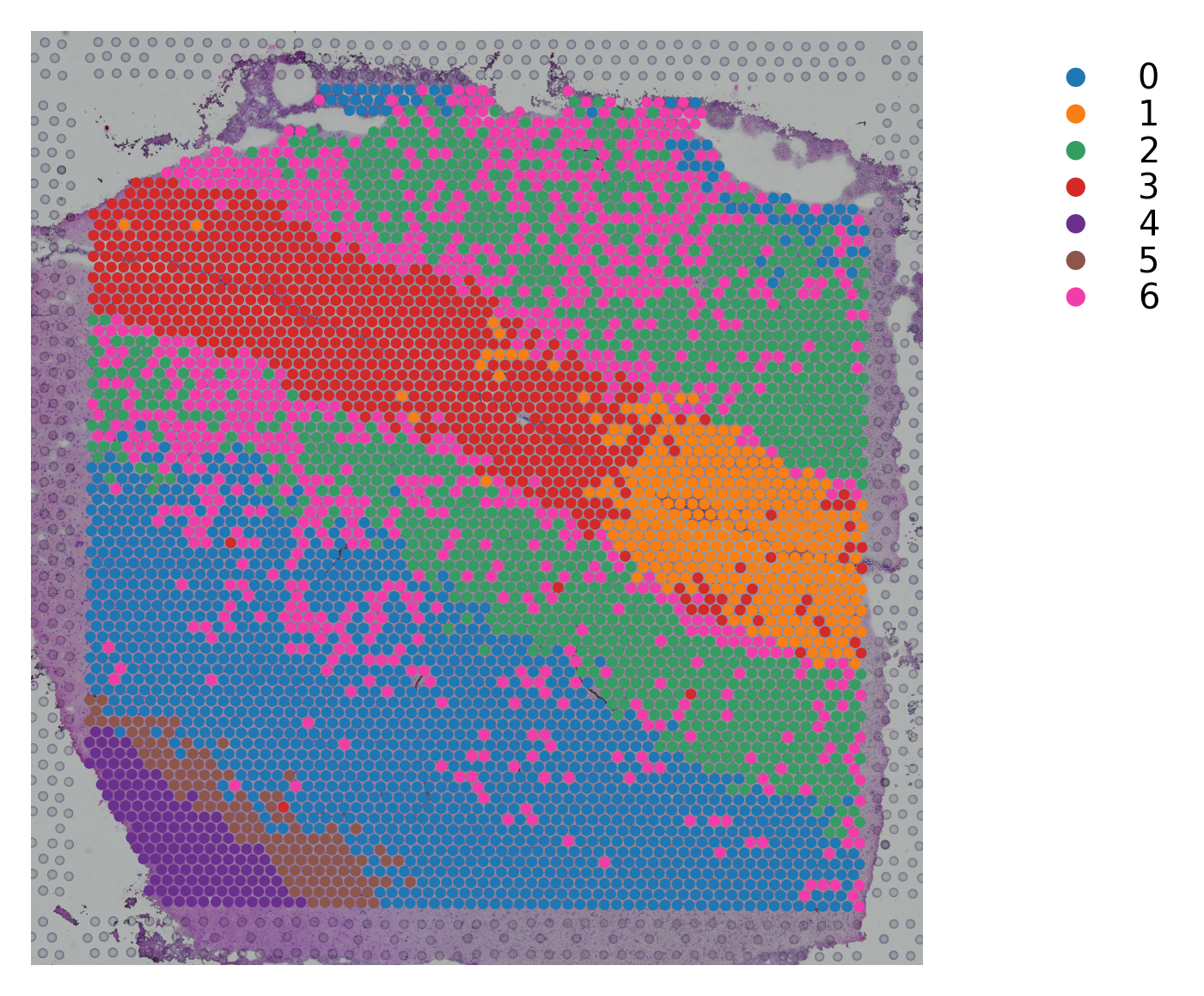
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
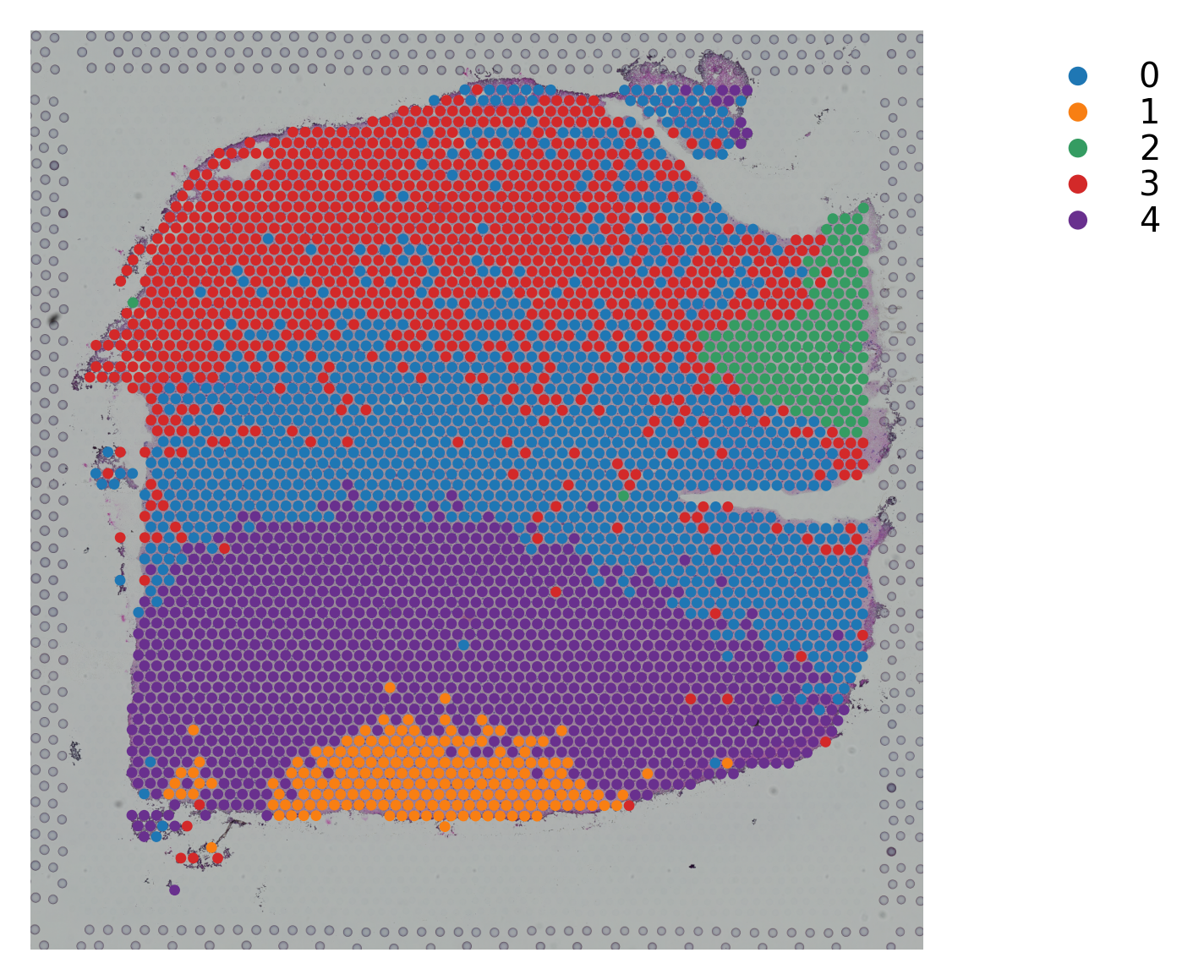
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
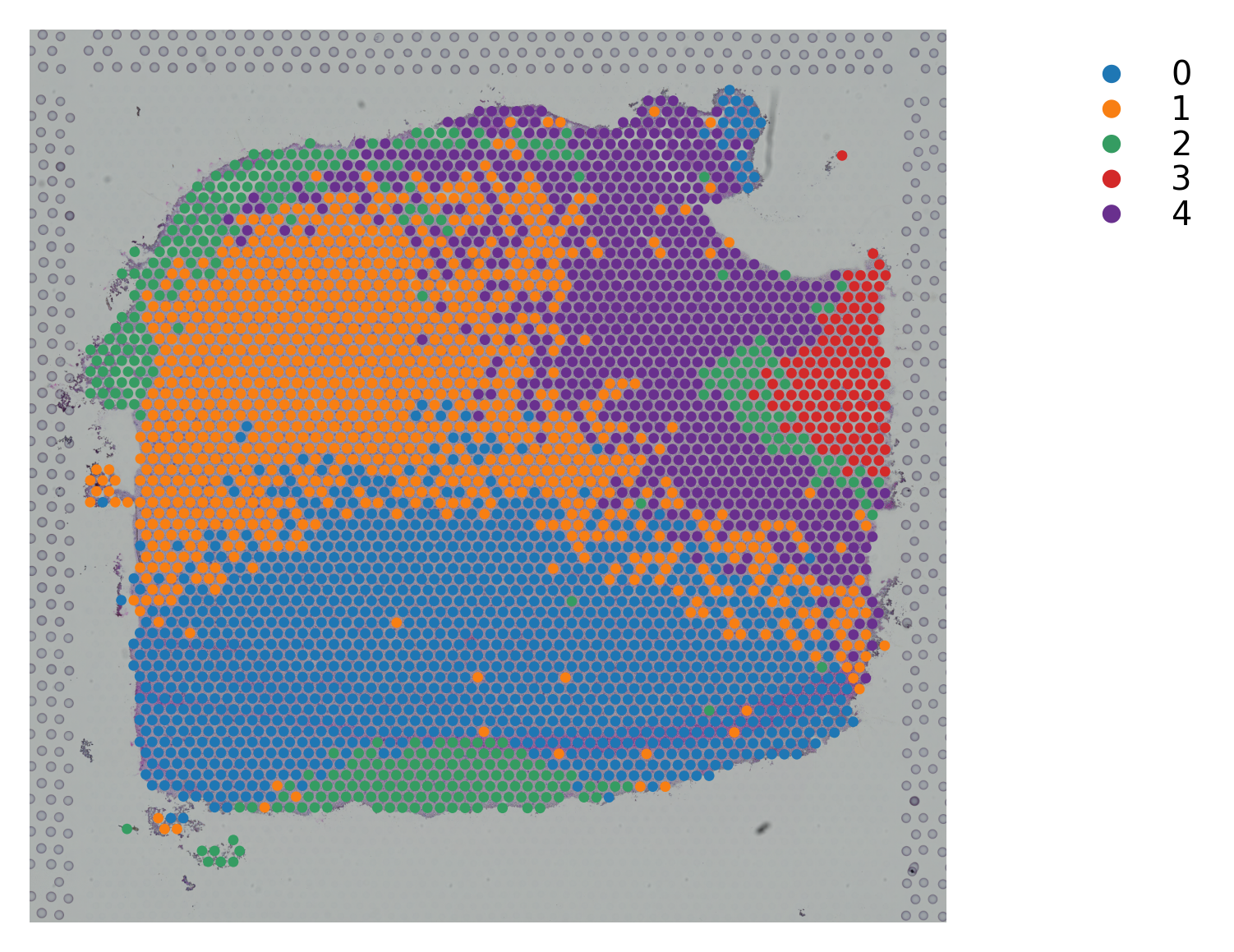
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
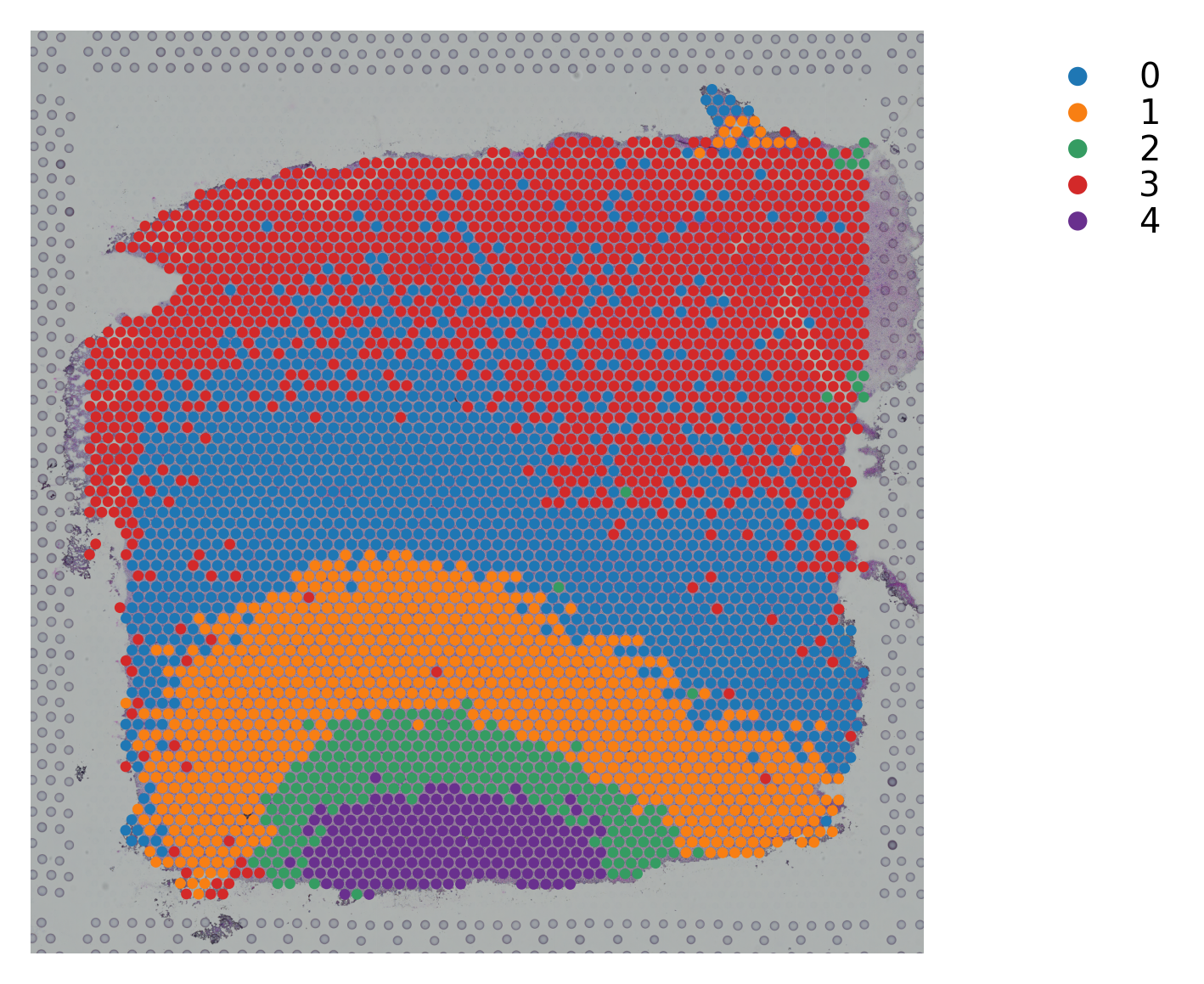
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:05 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
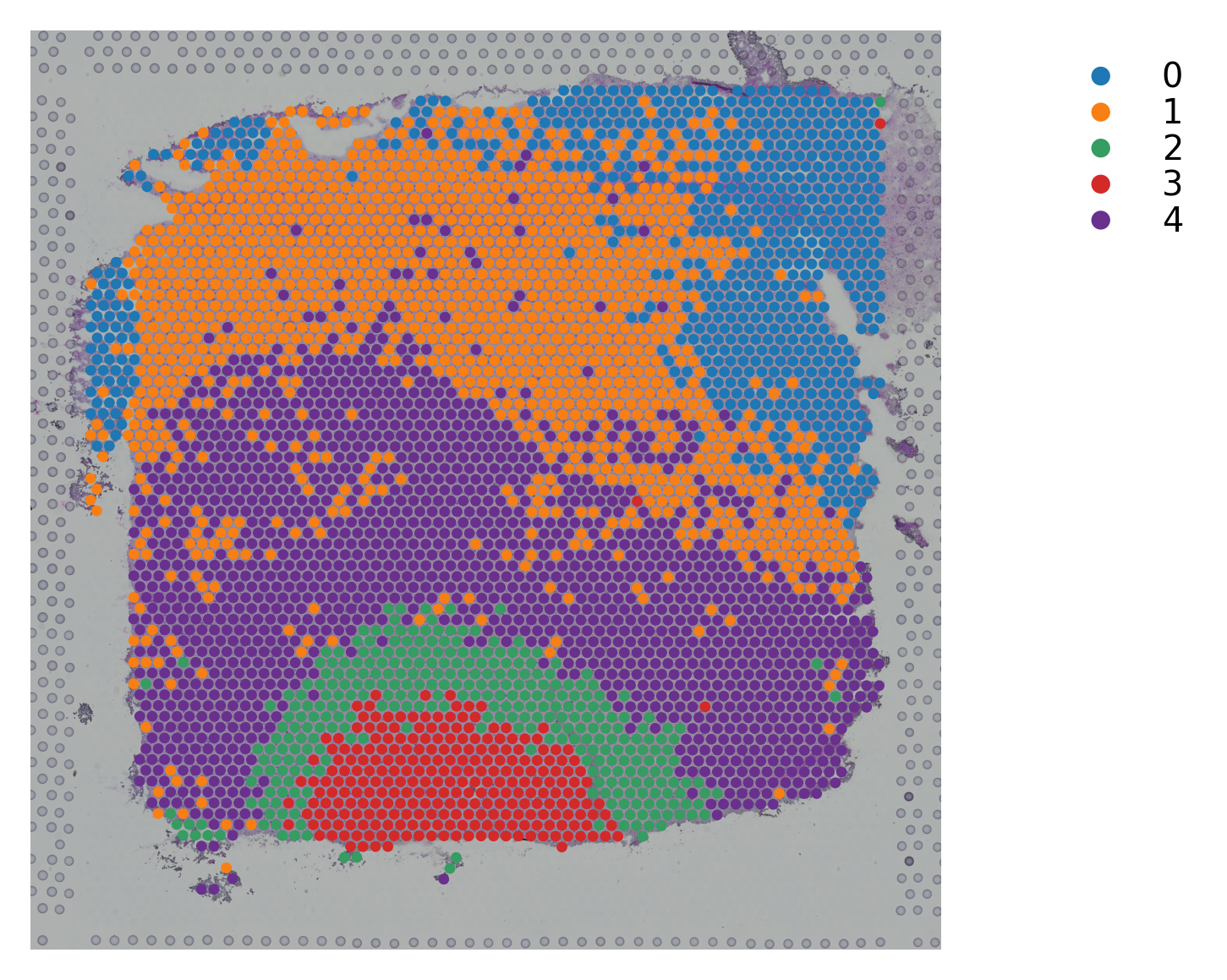
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
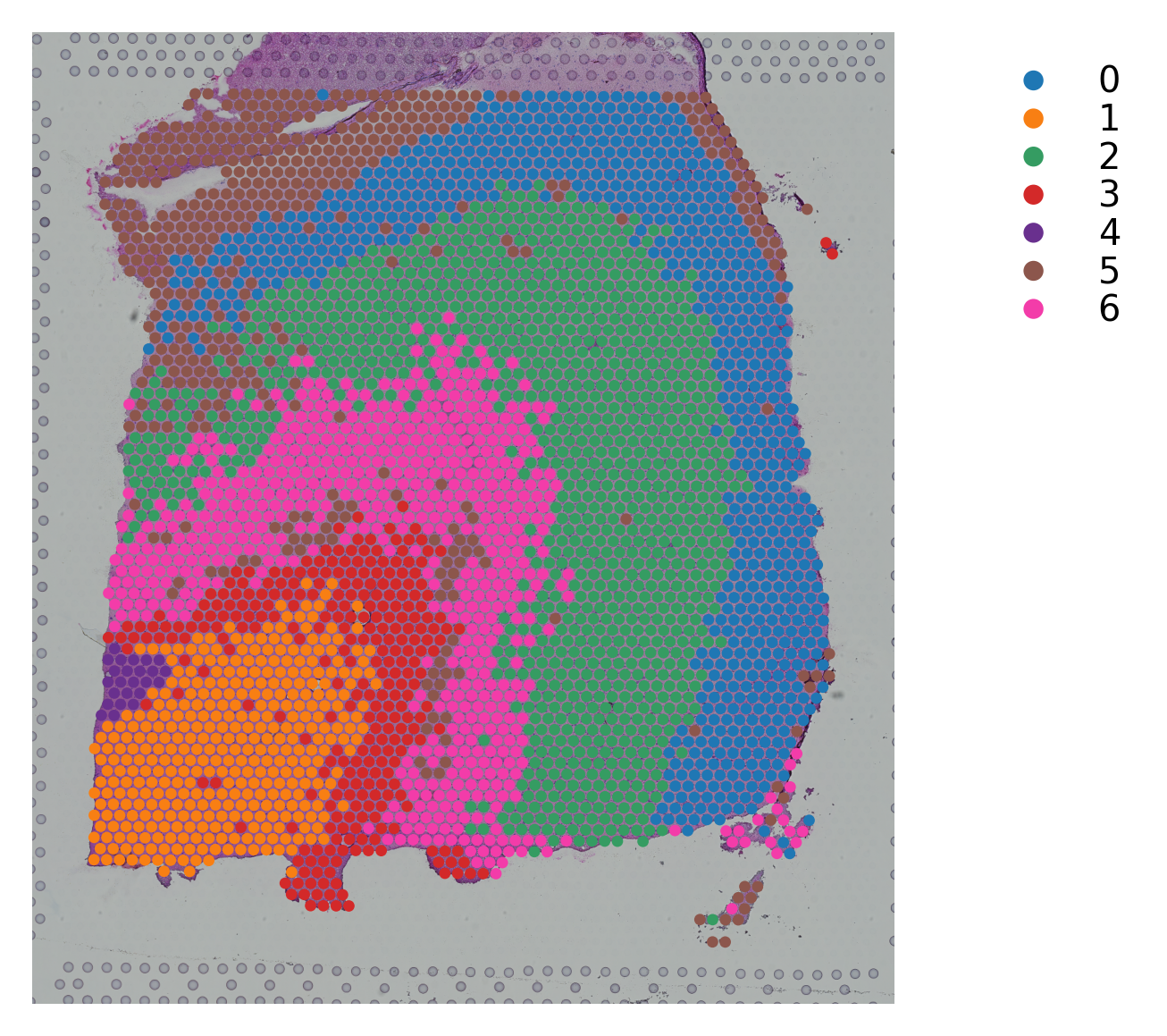
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
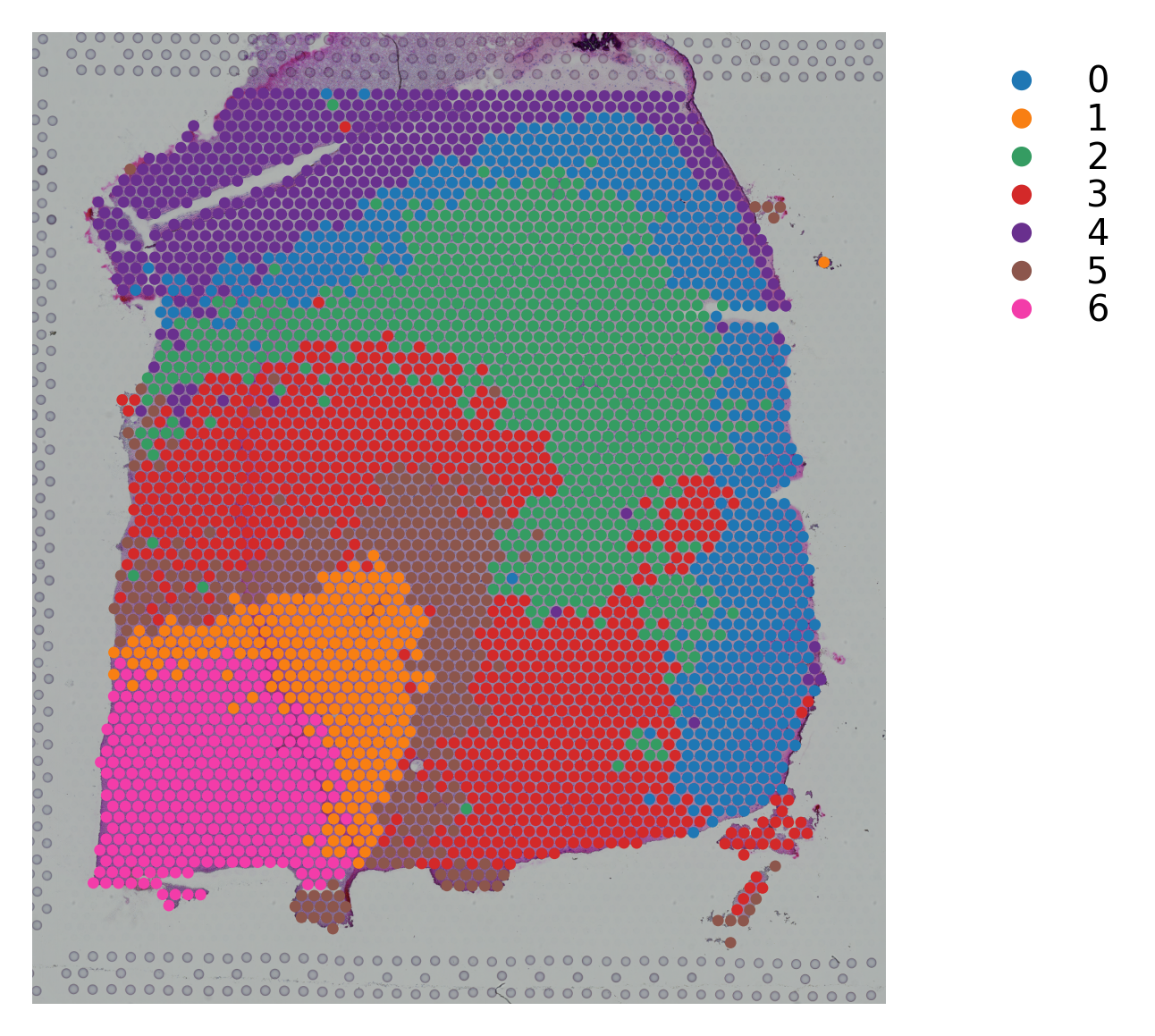
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
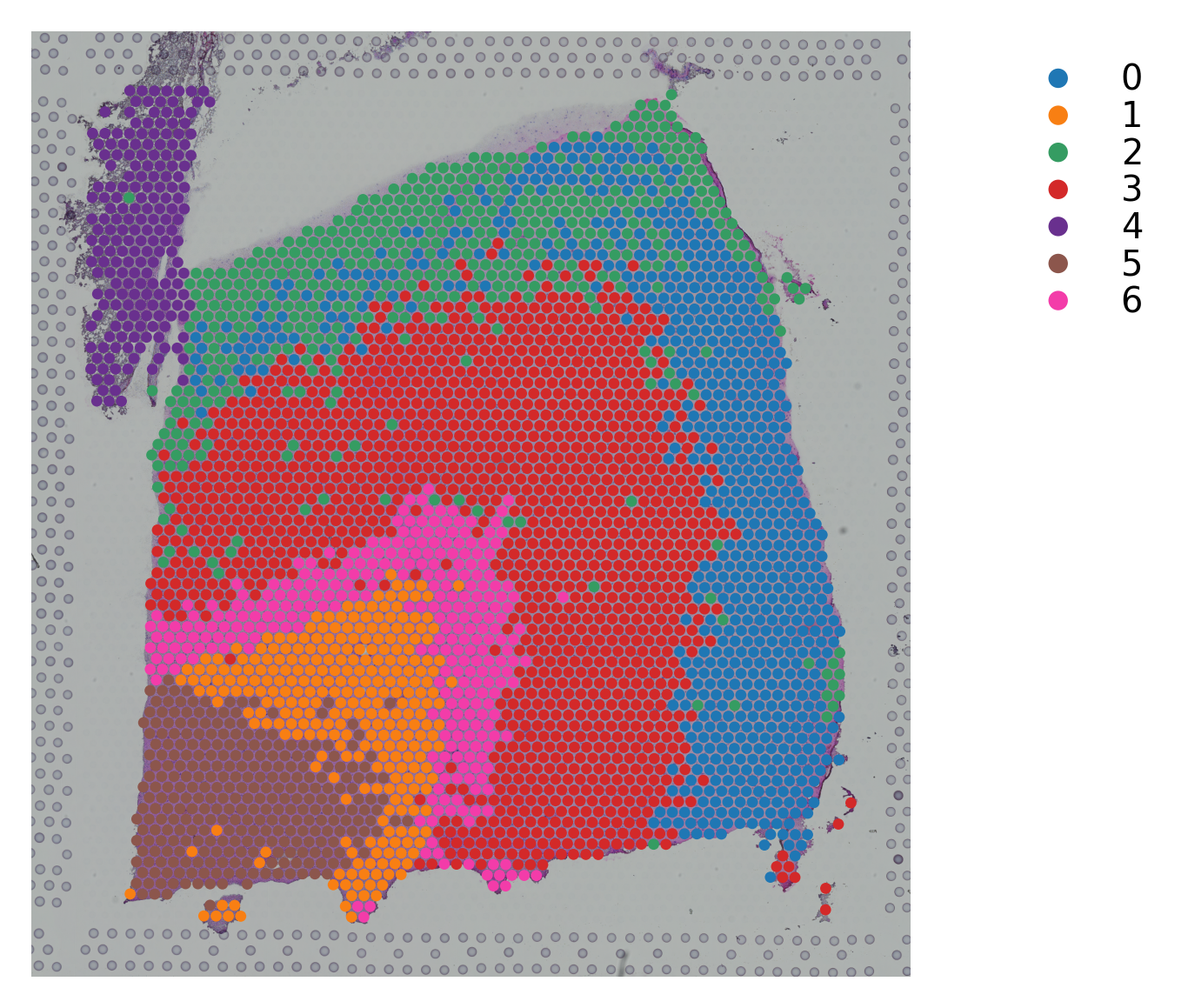
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Variable names are not unique. To make them unique, call `.var_names_make_unique`.
Normalization step is finished in adata.X
Log transformation step is finished in adata.X
Tiling image: 2%|▏ [ time left: 00:04 ]
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Tiling image: 100%|██████████ [ time left: 00:00 ]
Extract feature: 100%|██████████ [ time left: 00:00 ]
The morphology feature is added to adata.obsm['X_morphology']!
Adjusting data: 100%|██████████ [ time left: 00:00 ]
The data adjusted by SME is added to adata.obsm['raw_SME_normalized']
Scale step is finished in adata.X
PCA is done! Generated in adata.obsm['X_pca'], adata.uns['pca'] and adata.varm['PCs']
Applying Kmeans clustering ...
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:786: FutureWarning: 'precompute_distances' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25). It has no effect
warnings.warn("'precompute_distances' was deprecated in version "
/home/uqysun19/90days/.conda/envs/stlearn/lib/python3.8/site-packages/sklearn/cluster/_kmeans.py:792: FutureWarning: 'n_jobs' was deprecated in version 0.23 and will be removed in 1.0 (renaming of 0.25).
warnings.warn("'n_jobs' was deprecated in version 0.23 and will be"
Kmeans clustering is done! The labels are stored in adata.obs["kmeans"]
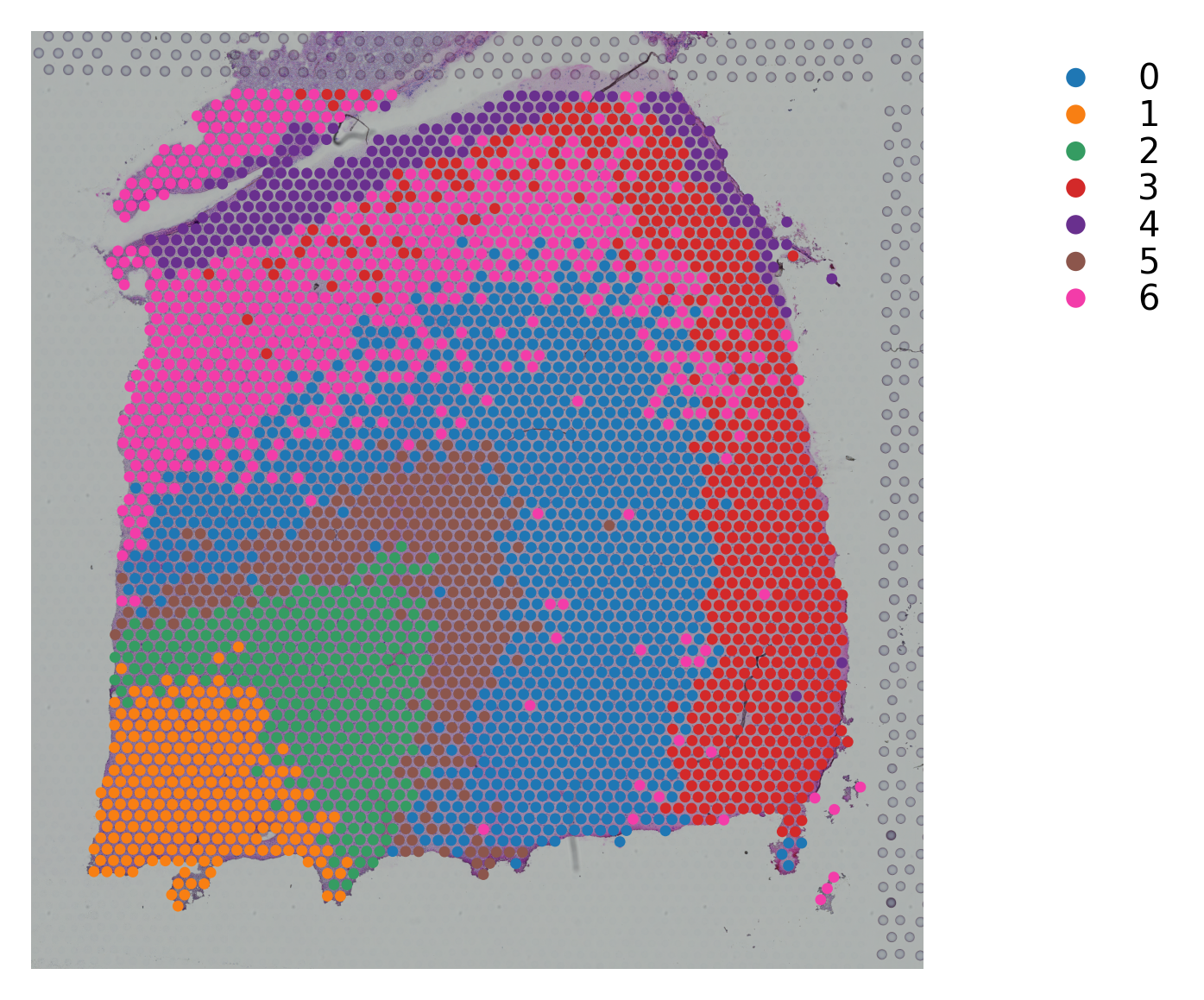
[26]:
# read clustering results from other methods
pca_df = pd.read_csv("./stSME_matrices_other_methods.csv")
[27]:
df_all = pca_df.append(df, ignore_index=True)
[28]:
import seaborn as sns
import matplotlib.pyplot as plt
sns.set_style("white")
from matplotlib.patches import PathPatch
def adjust_box_widths(g, fac):
"""
Adjust the widths of a seaborn-generated boxplot.
"""
# iterating through Axes instances
for ax in g.axes:
# iterating through axes artists:
for c in ax.get_children():
# searching for PathPatches
if isinstance(c, PathPatch):
# getting current width of box:
p = c.get_path()
verts = p.vertices
verts_sub = verts[:-1]
xmin = np.min(verts_sub[:, 0])
xmax = np.max(verts_sub[:, 0])
xmid = 0.5*(xmin+xmax)
xhalf = 0.5*(xmax - xmin)
# setting new width of box
xmin_new = xmid-fac*xhalf
xmax_new = xmid+fac*xhalf
verts_sub[verts_sub[:, 0] == xmin, 0] = xmin_new
verts_sub[verts_sub[:, 0] == xmax, 0] = xmax_new
# setting new width of median line
for l in ax.lines:
if np.all(l.get_xdata() == [xmin, xmax]):
l.set_xdata([xmin_new, xmax_new])
class GridShader():
def __init__(self, ax, first=True, **kwargs):
self.spans = []
self.sf = first
self.ax = ax
self.kw = kwargs
self.ax.autoscale(False, axis="x")
self.cid = self.ax.callbacks.connect('xlim_changed', self.shade)
self.shade()
def clear(self):
for span in self.spans:
try:
span.remove()
except:
pass
def shade(self, evt=None):
self.clear()
xticks = self.ax.get_xticks()
xlim = self.ax.get_xlim()
xticks = xticks[(xticks > xlim[0]) & (xticks < xlim[-1])]
locs = np.concatenate(([[xlim[0]], xticks+0.5, [xlim[-1]]]))
start = locs[1-int(self.sf)::2]
end = locs[2-int(self.sf)::2]
for s, e in zip(start, end):
self.spans.append(self.ax.axvspan(s, e, zorder=0, **self.kw))
[29]:
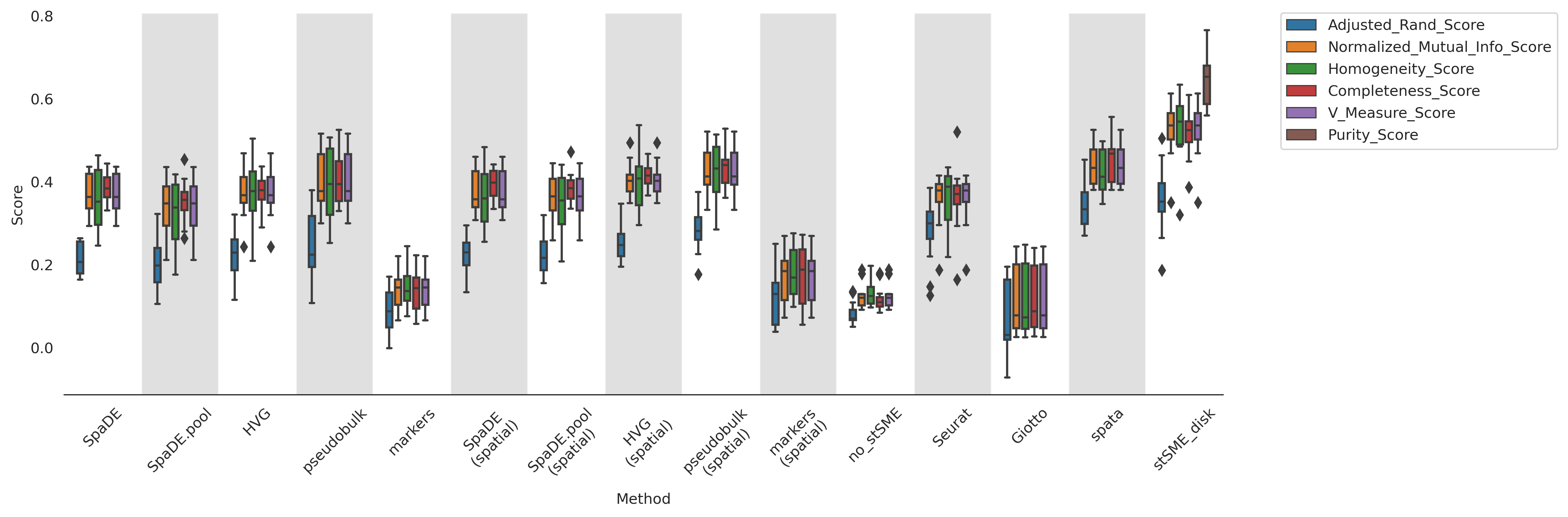
import seaborn as sns
fig = plt.figure(figsize=(15, 5))
a = sns.boxplot(x="Method", y="Score", hue="test",
width=0.7,
data=df_all)
a.set_xticklabels(a.get_xticklabels(), rotation=45)
sns.despine(left=True)
a.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
adjust_box_widths(fig, 0.7)
plt.autoscale()
gs = GridShader(a, facecolor="lightgrey", first=False, alpha=0.7)
plt.tight_layout()
#plt.savefig("./clustering_performace.png", dpi=300)
plt.show()
<ipython-input-28-1b10d13ee4c7>:36: DeprecationWarning: elementwise comparison failed; this will raise an error in the future.
if np.all(l.get_xdata() == [xmin, xmax]):